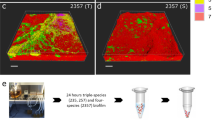
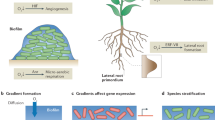
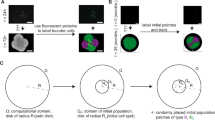
Key Points
-
Bacteria often exist in biofilms, which are surface-adhering or free-floating groups of cells that are bound together by a secreted polymer matrix. These microbial collectives are important for bacterial occupation of diverse ecological niches, they contribute to biogeochemical cycling, and they cause disease in multicellular organisms.
-
Within biofilms, bacteria interact with each other closely through cooperative phenotypes, such as the production of digestive enzymes, and antagonistic phenotypes, such as the expression of type V or type VI secretion systems. The evolutionary dynamics of these social phenotypes depend on their costs and their effects on other cells.
-
Many bacterial social phenotypes involve the secretion of products that affect neighbours in a distance-dependent manner. As a result, interaction networks within biofilms are largely determined by the spatial structure of the biofilms — that is, the arrangement in space of different clones, strains and species.
-
When biofilms are segregated into clonal clusters, the neighbourhood of a given cell mostly contains clonemates, and natural selection often favours the secretion of compounds that benefit all recipient cells. However, when different strains and species are spatially mixed within biofilms, cells primarily interact with other genotypes and antagonistic behaviour is often favoured. Under certain circumstances, between-species commensalism or mutualism can also evolve and remain stable against cheating.
-
Cooperative and antagonistic phenotypes fall under the control of sophisticated sensory mechanisms, such as competition sensing and quorum sensing, that evolved to help account for the variation in exposure to other strains and species in space and time. These regulatory mechanisms help to reduce the marginal costs of social phenotypes, maximize their fitness impacts and ensure that the correct recipient cells are targeted.
-
Both cooperative and antagonistic behaviours feed back onto population spatial structure by locally altering the growth rates of other cells and thus changing local biofilm composition.
-
Many bacteria and unicellular eukaryotes have evolved strategies for actively altering biofilm population structure, either through selective adhesion that spatially assorts the biofilm into groups that contain one or more specific genotypes or through the secretion of extracellular matrix components that spatially organize biofilm-dwelling cells.
Abstract
Bacteria often live within matrix-embedded communities, termed biofilms, which are now understood to be a major mode of microbial life. The study of biofilms has revealed their vast complexity both in terms of resident species composition and phenotypic diversity. Despite this complexity, theoretical and experimental work in the past decade has identified common principles for understanding microbial biofilms. In this Review, we discuss how the spatial arrangement of genotypes within a community influences the cooperative and competitive cell–cell interactions that define biofilm form and function. Furthermore, we argue that a perspective rooted in ecology and evolution is fundamental to progress in microbiology.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Hall-Stoodley, L., Costerton, J. W. & Stoodley, P. Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2, 95–108 (2004).
Hobley, L., Harkins, C., MacPhee, C. E. & Stanley-Wall, N. R. Giving structure to the biofilm matrix: an overview of individual strategies and emerging common themes. FEMS Microbiol. Rev. 39, 649–669 (2015).
Arnosti, C. Microbial extracellular enzymes and the marine carbon cycle. Annu. Rev. Mar. Sci. 3, 401–425 (2011).
Battin, T. J., Kaplan, L. A., Newbold, J. D. & Hansen, C. M. E. Contributions of microbial biofilms to ecosystem processes in stream mesocosms. Nature 426, 439–442 (2003).
Macfarlane, S., Bahrami, B. & Macfarlane, G. T. Mucosal biofilm communities in the human intestinal tract. Adv. Appl. Microbiol. 75, 111–143 (2011).
Hoiby, N., Bjarnsholt, T., Givskov, M., Molin, S. & Ciofu, O. Antibiotic resistance of bacterial biofilms. Int. J. Antimicrob. Agents 35, 322–332 (2010).
Bixler, G. D. & Bhushan, B. Biofouling: lessons from nature. Philos. Trans. A Math. Phys. Eng. Sci. 370, 2381–2417 (2012).
Drescher, K., Shen, Y., Bassler, B. L. & Stone, H. A. Biofilm streamers cause catastrophic disruption of flow with consequences for environmental and medical systems. Proc. Natl Acad. Sci. USA 110, 4345–4350 (2013).
Harding, J. L. & Reynolds, M. M. Combating medical device fouling. Trends Biotechnol. 32, 140–146 (2014).
Nadell, C. D. et al. Cutting through the complexity of cell collectives. Proc. Biol. Sci. 280, 20122770 (2013).
Nadell, C. D., Xavier, J. B. & Foster, K. R. The sociobiology of biofilms. FEMS Microbiol. Rev. 33, 206–224 (2009).
Visca, P., Imperi, F. & Lamont, I. Pyoverdine siderophores: from biogenesis to biosignificance. Trends Microbiol. 15, 22–30 (2007).
Griffin, A. S., West, S. A. & Buckling, A. Cooperation and competition in pathogenic bacteria. Nature 430, 1024–1027 (2004). A key proof-of-principle investigation finding that secreted siderophores can act as public goods that are susceptible to the evolution of cheating behaviour.
Allison, S. D. Cheaters, diffusion and nutrients constrain decomposition by microbial enzymes in spatially structured environments. Ecol. Lett. 8, 626–635 (2005).
Absalon, C., Van Dellen, K. & Watnick, P. I. A communal bacterial adhesin anchors biofilm and bystander cells to surfaces. PLoS Pathog. 7, e1002210 (2011).
Xavier, J. B., Kim, W. & Foster, K. R. A molecular mechanism that stabilizes cooperative secretions in Pseudomonas aeruginosa. Mol. Microbiol. 79, 166–179 (2011).
Flemming, H.-C. & Wingender, J. The biofilm matrix. Nat. Rev. Microbiol. 8, 623–633 (2010).
West, S. A., Diggle, S. P., Buckling, A., Gardner, A. & Griffins, A. S. The social lives of microbes. Annu. Rev. Ecol. Evol. Syst. 38, 53–77 (2007).
Meibom, K. L. et al. The Vibrio cholerae chitin utilization program. Proc. Natl Acad. Sci. USA 101, 2524–2529 (2004).
Drescher, K., Nadell, C., Stone, H., Wingreen, N. & Bassler, B. Solutions to the public goods dilemma in bacterial biofilms. Curr. Biol. 24, 50–55 (2014).
Cordero, O. X., Ventouras, L. A., DeLong, E. F. & Polz, M. F. Public good dynamics drive evolution of iron acquisition strategies in natural bacterioplankton populations. Proc. Natl Acad. Sci. USA 109, 20059–20064 (2012).
Matz, C. et al. Biofilm formation and phenotypic variation enhance predation-driven persistence of Vibrio cholerae. Proc. Natl Acad. Sci. USA 102, 16819–16824 (2005).
Hibbing, M. E., Fuqua, C., Parsek, M. R. & Peterson, S. B. Bacterial competition: surviving and thriving in the microbial jungle. Nat. Rev. Microbiol. 8, 15–25 (2010).
Rendueles, O. & Ghigo, J. M. Mechanisms of competition in biofilm communities. Microbiol. Spectr. 3, 3 (2015).
Riley, M. A. & Wertz, J. E. Bacteriocins: evolution, ecology, and application. Annu. Rev. Microbiol. 56, 117–137 (2002).
Hayes, C. S., Aoki, S. K. & Low, D. A. Bacterial contact-dependent delivery systems. Annu. Rev. Genet. 44, 71–90 (2010).
Ho, B. T., Dong, T. G. & Mekalanos, J. J. A view to a kill: the bacterial type VI secretion system. Cell Host Microbe. 15, 9–21 (2014).
Russell, A. B., Peterson, S. B. & Mougous, J. D. Type VI secretion system effectors: poisons with a purpose. Nat. Rev. Microbiol. 12, 137–148 (2014).
Basler, M., Ho, B. T. & Mekalanos, J. J. Tit-for-tat: type VI secretion system counterattack during bacterial cell-cell interactions. Cell 152, 884–894 (2013). A report showing that the T6SS of P. aeruginosa is deployed in response to the T6SS-mediated attack from other species in the vicinity.
Nadell, C. D. & Bassler, B. L. A fitness trade-off between local competition and dispersal in Vibrio cholerae biofilms. Proc. Natl Acad. Sci. USA 108, 14181–14185 (2011).
Schluter, J., Nadell, C. D., Bassler, B. L. & Foster, K. R. Adhesion as a weapon in microbial competition. ISME J. 9, 139–149 (2015).
Kim, W., Racimo, F., Schluter, J., Levy, S. B. & Foster, K. R. Importance of positioning for microbial evolution. Proc. Natl Acad. Sci. USA 111, E1639–E1647 (2014).
Rumbaugh, K. P. et al. Quorum sensing and the social evolution of bacterial virulence. Curr. Biol. 19, 341–345 (2009). A study demonstrating that phenotypes that are regulated by quorum sensing can be exploited by cheating mutants within a population of P. aeruginosa during infection of a mouse model system.
Inglis, R. F., Gardner, A., Cornelis, P. & Buckling, A. Spite and virulence in the bacterium Pseudomonas aeruginosa. Proc. Natl Acad. Sci. USA 106, 5703–5707 (2009).
Brown, S. P., Inglis, R. F. & Taddei, F. Evolutionary ecology of microbial wars: within-host competition and (incidental) virulence. Evol. Appl. 2, 32–39 (2009).
Levin, S. A. Complex adaptive systems: exploring the known, the unknown, and the unknowable. Bull. Am. Math. Soc. 40, 3–19 (2003).
Persat, A. et al. The mechanical world of bacteria. Cell 161, 988–997 (2015).
Stacy, A., McNally, L., Darch, S., Brown, S. P. & Whiteley, M. The biogeography of polymicrobial infection. Nat. Rev. Microbiol. 14, 93–105 (2015). A major recent review of processes that generate the spatial structure of different bacterial strains and species in microbial communities associated with infection.
Driscoll, W. W. & Pepper, J. W. Theory for the evolution of diffusible external goods. Evolution 64, 2682–2687 (2010).
Lion, S. & van Baalen, M. Self-structuring in spatial evolutionary ecology. Ecol. Lett. 11, 277–295 (2008).
O'Toole, G. A. & Wong, G. C. Sensational biofilms: surface sensing in bacteria. Curr. Opin. Microbiol. 30, 139–146 (2016).
Millet, Y. A. et al. Insights into Vibrio cholerae intestinal colonization from monitoring fluorescently labeled bacteria. PLoS Pathog. 10, e1004405 (2014).
Anderson, M. S., Garcia, E. C. & Cotter, P. A. Kind discrimination and competitive exclusion mediated by contact-dependent growth inhibition systems shape biofilm community structure. PLoS Pathog. 10, e1004076 (2014).
Nadell, C. D., Foster, K. R. & Xavier, J. B. Emergence of spatial structure in cell groups and the evolution of cooperation. PLoS Comput. Biol. 6, e1000716 (2010).
Thomas, C. D. & Kunin, W. E. The spatial structure of populations. J. Animal Ecol. 68, 647–657 (1999).
Hallatschek, O., Hersen, P., Ramanathan, S. & Nelson, D. R. Genetic drift at expanding frontiers promotes gene segregation. Proc. Natl Acad. Sci. USA 104, 19926–19930 (2007). A theoretical and experimental paper that outlines how spatial structure emerges along the leading edge of expanding bacterial colonies owing to genetic drift, which generates clonal patches of one genotype.
Weber, M. F., Poxleitner, G., Hebisch, E., Frey, E. & Opitz, M. Chemical warfare and survival strategies in bacterial range expansions. J. R. Soc. Interface 11, 20140172 (2014).
van Gestel, J., Weissing, F. J., Kuipers, O. P. & Kovacs, A. T. Density of founder cells affects spatial pattern formation and cooperation in Bacillus subtilis biofilms. ISME J. 8, 2069–2079 (2014).
Mitri, S., Clarke, E. & Foster, K. R. Resource limitation drives spatial organization in microbial groups. ISME J. 10, 1471–1482 (2016).
Van Dyken, J. D., Muller, M. J. I., Mack, K. M. L. & Desai, M. M. Spatial population expansion promotes the evolution of cooperation in an experimental prisoner's dilemma. Curr. Biol. 23, 919–923 (2013).
Müller, M., Neugeboren, B. I., Nelson, D. R. & Murray, A. W. Genetic drift opposes mutualism during spatial population expansion. Proc. Natl Acad. Sci. USA 111, 1037–1042 (2014). Research demonstrating that genetic drift in expanding S. cerevisiae colonies generates a spatial structure that inhibits cooperation between two genotypes in a synthetic system. By contrast, strong mutualism is shown to counteract the lineage-segregating influence of radial population growth.
Buttery, N. et al. Structured growth and genetic drift raise relatedness in the social amoeba Dictyostelium discoideum. Biol. Lett. 8, 794–797 (2012).
Poilane, I., Karjalainen, T., Barc, M.-C., Bourlioux, P. & Collignon, A. Protease activity of Clostridium difficile strains. Can. J. Microbiol. 44, 157–161 (1998).
Hungate, R. The anaerobic mesophilic cellulolytic bacteria. Bacteriol. Rev. 14, 1 (1950).
Gilbert, H. J. & Hazlewood, G. P. Bacterial cellulases and xylanases. Microbiology 139, 187–194 (1993).
Ross-Gillespie, A., Gardner, A., West, S. A. & Griffin, A. S. Frequency dependence and cooperation: theory and a test with bacteria. Am. Nat. 170, 331–342 (2007).
Kümmerli, R., Schiessl, K. T., Waldvogel, T., McNeill, K. & Ackermann, M. Habitat structure and the evolution of diffusible siderophores in bacteria. Ecol. Lett. 17, 1536–1544 (2014).
West, S. A., Griffin, A. S., Gardner, A. & Diggle, S. P. Social evolution theory for microorganisms. Nat. Rev. Microbiol. 4, 597–607 (2006).
Köhler, T., Buckling, A. & van Delden, C. Cooperation and virulence of clinical Pseudomonas aeruginosa populations. Proc. Natl Acad. Sci. USA 106, 6339–6344 (2009).
Andersen, S. B. et al. Long-term social dynamics drive loss of function in pathogenic bacteria. Proc. Natl Acad. Sci. USA 112, 10756–10761 (2015).
Allen, B., Gore, J. & Nowak, M. A. Spatial dilemmas of diffusible public goods. eLife 2, e01169 (2013).
Borenstein, D. B., Meir, Y., Shaevitz, J. W. & Wingreen, N. S. Non-local interaction via diffusible resource prevents coexistence of cooperators and cheaters in a lattice model. PLoS ONE 8, e63304 (2013).
Damore, J. A. & Gore, J. Understanding microbial cooperation. J. Theor. Biol. 299, 31–41 (2012).
Dobay, A., Bagheri, H. C., Messina, A., Kümmerli, R. & Rankin, D. J. Interaction effects of cell diffusion, cell density and public goods properties on the evolution of cooperation in digital microbes. J. Evol. Biol. 27, 1869–1877 (2014).
Popat, R. et al. Quorum-sensing and cheating in bacterial biofilms. Proc. Biol. Sci. 279, 4765–4771 (2012).
Hamilton, W. D. The genetical evolution of social behaviour I. J. Theor. Biol. 7, 1–16 (1964).
Hamilton, W. D. The genetical evolution of social behaviour II. J. Theor. Biol. 7, 17–52 (1964). Landmark papers in evolutionary biology, establishing the fundamental theory and broad-ranging importance of genetic identity between individuals for the evolution of cooperation.
Mitri, S., Xavier, J. B. & Foster, K. R. Social evolution in multispecies biofilms. Proc. Natl Acad. Sci. USA 108, 10839–10846 (2011).
Kümmerli, R., Griffin, A. S., West, S. A., Buckling, A. & Harrison, F. Viscous medium promotes cooperation in the pathogenic bacterium Pseudomonas aeruginosa. Proc. Biol. Sci. 276, 3531–3538 (2009).
Julou, T. et al. Cell–cell contacts confine public goods diffusion inside Pseudomonas aeruginosa clonal microcolonies. Proc. Natl Acad. Sci. USA 110, 12577–12582 (2013).
Seminara, A. et al. Osmotic spreading of Bacillus subtilis biofilms driven by an extracellular matrix. Proc. Natl Acad. Sci. USA 109, 1116–1121 (2012).
Datta, M. S., Korolev, K. S., Cvijovic, I., Dudley, C. & Gore, J. Range expansion promotes cooperation in an experimental microbial metapopulation. Proc. Natl Acad. Sci. USA 110, 7354–7359 (2013). This paper and reference 50 provide evidence that genetic drift in expanding metapopulations of S. cerevisiae generates a spatial structure which favours the use of a cooperative enzyme by a single genotype.
Korolev, K. S., Xavier, J. B., Nelson, D. R. & Foster, K. R. A. Quantitative test of population genetics using spatiogenetic patterns in bacterial colonies. Am. Nat. 178, 538–552 (2011).
Hol, F. J. H. et al. Spatial structure facilitates cooperation in a social dilemma: empirical evidence from a bacterial community. PLoS ONE 8, e77042 (2013).
Mitri, S. & Foster, K. R. The genotypic view of social interactions in microbial communities. Annu. Rev. Genet. 47, 247–273 (2013).
Foster, K. R. & Bell, T. Competition, not cooperation, dominates interactions among culturable microbial species. Curr. Biol. 22, 1845–1850 (2012).
Oliveria, N. M. et al. Biofilm formation as a response to ecological competition. PLoS Biol. 13, e1002191 (2015).
Pfeiffer, T. Cooperation and competition in the evolution of ATP-producing pathways. Science 292, 504–507 (2001).
Xavier, J. B. & Foster, K. R. Cooperation and conflict in microbial biofilms. Proc. Natl Acad. Sci. USA 104, 876–881 (2007).
Durrett, R. & Levin, S. Allelopathy in spatially distributed populations. J. Theor. Biol. 185, 165–171 (1997).
Ratcliff, W. & Denison, R. Alternative actions for antibiotics. Science 332, 547–548 (2011).
Abrudan, M. I. et al. Socially mediated induction and suppression of antibiosis during bacterial coexistence. Proc. Natl Acad. Sci. USA 112, 11054–11059 (2015).
Borgeaud, S., Metzger, L. C., Scrignari, T. & Blokesch, M. The type VI secretion system of Vibrio cholerae fosters horizontal gene transfer. Science 347, 63–67 (2015).
Gardner, A. & West, S. A. Spite and the scale of competition. J. Evol. Biol. 17, 1195–1203 (2004).
Bucci, V., Nadell, C. D. & Xavier, J. B. The evolution of bacteriocin production in bacterial biofilms. Am. Nat. 178, E162–E173 (2011).
Tait, K. & Sutherland, I. W. Antagonistic interactions amongst bacteriocin-producing enteric bacteria in dual species biofilms. J. Appl. Microbiol. 93, 345–352 (2002).
Borenstein, D. B., Ringel, P., Basler, M. & Wingreen, N. S. Established microbial colonies can survive type VI secretion assault. PLoS Comput. Biol. 11, e1004520 (2015).
Wexler, A. G. et al. Human symbionts inject and neutralize antibacterial toxins to persist in the gut. Proc. Natl Acad. Sci. USA 113, 3639–3644 (2016).
Alteri, C. J. et al. Multicellular bacteria deploy the type VI secretion system to preemptively strike neighboring cells. PLoS Pathog. 9, e1003608 (2013).
Karlsson, F. H., Nookaew, I., Petranovic, D. & Nielsen, J. Prospects for systems biology and modeling of the gut microbiome. Trends Biotechnol. 29, 251–258 (2011).
Morris, J. J., Lenski, R. E. & Zinser, E. R. The black queen hypothesis: evolution of dependencies through adaptive gene loss. mBio 3, e00036-12 (2012).
Tripp, H. J. et al. SAR11 marine bacteria require exogenous reduced sulphur for growth. Nature 452, 741–744 (2008).
Oliveira, N. M., Niehus, R. & Foster, K. R. Evolutionary limits to cooperation in microbial communities. Proc. Natl Acad. Sci. USA 111, 17941–17946 (2014).
Estrela, S. & Brown, S. P. Metabolic and demographic feedbacks shape the emergent spatial structure and function of microbial communities. PLoS Comput. Biol. 9, e1003398 (2013).
Momeni, B., Waite, A. J. & Shou, W. Spatial self-organization favors heterotypic cooperation over cheating. eLife 2, e00960 (2013). A study in which synthetic obligate mutualist strains of S. cerevisiae are found to spatially exclude a cheating strain in surface-bound colonies in a manner that promotes cooperation between mutualists.
Morris, B. E. L., Henneberger, R., Huber, H. & Moissl-Eichinger, C. Microbial syntrophy: interaction for the common good. FEMS Microbiol. Rev. 37, 384–406 (2013).
Callaghan, A. et al. The genome sequence of Desulfatibacillum alkenivorans AK-01: a blueprint for anaerobic alkane oxidation. Environ. Microbiol. 14, 101–113 (2012).
Schink, B. Synergistic interactions in the microbial world. Antonie Van Leeuwenhoek 81, 257–261 (2002).
Pande, S. et al. Fitness and stability of obligate cross-feeding interactions that emerge upon gene loss in bacteria. ISME J. 8, 953–962 (2014).
Rakoff-Nahoum, S., Foster, K. R. & Comstock, L. The evolution of cooperation within the gut microbiota. Nature 533, 255–259 (2016).
Estrela, S., Trisos, C. H. & Brown, S. P. From metabolism to ecology: cross-feeding interactions shape the balance between polymicrobial conflict and mutualism. Am. Nat. 180, 566–576 (2012).
Momeni, B., Brileya, K. A., Fields, M. W. & Shou, W. Strong inter-population cooperation leads to partner intermixing in microbial communities. eLife 2, e00230 (2013).
Kümmerli, R., Jiricny, N., Clarke, L. S., West, S. A. & Griffin, A. S. Phenotypic plasticity of a cooperative behaviour in bacteria. J. Evol. Biol. 22, 589–598 (2009).
Kümmerli, R. & Brown, S. P. Molecular and regulatory properties of a public good shape the evolution of cooperation. Proc. Natl Acad. Sci. USA 107, 18921–18926 (2010).
Brown, S. P. & Taddei, F. The durability of public goods changes the dynamics and nature of social dilemmas. PLoS ONE 2, e593 (2007).
Mellbye, B. & Schuster, M. Physiological framework for the regulation of quorum sensing-dependent public goods in Pseudomonas aeruginosa. J. Bacteriol. 196, 1155–1164 (2014).
Cornforth, D. M. & Foster, K. R. Competition sensing: the social side of bacterial stress responses. Nat. Rev. Microbiol. 11, 285–293 (2013).
Schuster, M., Sexton, D. J., Diggle, S. P. & Greenberg, E. P. Acyl-homoserine lactone quorum sensing: from evolution to application. Annu. Rev. Microbiol. 67, 43–63 (2013).
Ng, W.-L. & Bassler, B. L. Bacterial quorum-sensing network architectures. Annu. Rev. Genet. 43, 197–222 (2009).
Redfield, R. J. Is quorum sensing a side effect of diffusion sensing? Trends Microbiol. 10, 365–370 (2002).
Cornforth, D. M. et al. Combinatorial quorum sensing allows bacteria to resolve their social and physical environment. Proc. Natl Acad. Sci. USA 111, 4280–4284 (2014).
Kim, M. K., Ingremeau, F., Zhao, A., Bassler, B. L. & Stone, H. A. Local and global consequences of flow on bacterial quorum sensing. Nat. Microbiol. 1, 15005 (2016).
Nadell, C. D., Xavier, J. B., Levin, S. A. & Foster, K. R. The evolution of quorum sensing in bacterial biofilms. PLoS Biol. 6, e14 (2008).
Schluter, J., Schoech, A., Foster, K. R. & Mitri, S. The evolution of quorum sensing as a mechanism to infer kinship. PLoS Comput. Biol. 12, e1004848 (2016).
van der Ploeg, J. R. Regulation of bacteriocin production in Streptococcus mutans by the quorum-sensing system required for development of genetic competence. J. Bacteriol. 187, 3980–3989 (2005).
Fontaine, L. et al. Quorum-sensing regulation of the production of Blp bacteriocins in Streptococcus thermophilus. J. Bacteriol. 189, 7195–7205 (2007).
Risøen, P. A., Brurberg, M. B., Eijsink, V. G. & Nes, I. F. Functional analysis of promoters involved in quorum sensing-based regulation of bacteriocin production in Lactobacillus. Mol. Microbiol. 37, 619–628 (2000).
LeRoux, M., Peterson, S. B. & Mougous, J. D. Bacterial danger sensing. J. Mol. Biol. 427, 3744–3753 (2015).
Korgaonkar, A. K. & Whiteley, M. Pseudomonas aeruginosa enhances production of an antimicrobial in response to N-acetylglucosamine and peptidoglycan. J. Bacteriol. 193, 909–917 (2011).
Dong, T. G. et al. Generation of reactive oxygen species by lethal attacks from competing microbes. Proc. Natl Acad. Sci. USA 112, 2181–2186 (2015).
LeRoux, M. et al. Kin cell lysis is a danger signal that activates antibacterial pathways of Pseudomonas aeruginosa. eLife 4, e05701 (2015). An investigation showing that cell lysate upregulates the T6SS of P. aeruginosa such that cells attack when they detect cues of clonemate death in the near surroundings.
Nakamaru, M., Matsuda, H. & Iwasa, Y. The evolution of cooperation in a lattice-structured population. J. Theor. Biol. 184, 65–81 (1997).
Durrett, R. & Levin, S. The importance of being discrete (and spatial). Theor. Popul. Biol. 46, 363–394 (1994).
Mitteldorf, J. & Wilson, D. S. Population viscosity and the evolution of altruism. J. Theor. Biol. 204, 481–496 (2000).
Ratzke, C. & Gore, J. Self-organized patchiness facilitates survival in cooperatively growing Bacillus subtilis populations. Nat. Microbiol. 1, 16022 (2016).
Hallatschek, O. & Nelson, D. R. Gene surfing in expanding populations. Theor. Popul. Biol. 73, 158–170 (2008).
Kerr, B., Riley, M. A., Feldman, M. W. & Bohannan, B. J. M. Local dispersal promotes biodiversity in a real-life game of rock-paper-scissors. Nature 418, 171–174 (2002).
Pande, S. et al. Privatization of cooperative benefits stabilizes mutualistic cross-feeding interactions in spatially structured environments. ISME J. 10, 1413–1423 (2016).
Tolker-Nielsen, T. & Molin, S. Spatial organization of microbial biofilm communities. Microb. Ecol. 40, 75–84 (2000).
Rendueles, O. et al. Rapid and widespread de novo evolution of kin discrimination. Proc. Natl Acad. Sci. USA 112, 9076–9081 (2015).
Strassmann, J. E., Gilbert, O. M. & Queller, D. C. Kin discrimination and cooperation in microbes. Annu. Rev. Microbiol. 65, 349–367 (2011).
Oldewurtel, E. R., Kouzel, N., Dewenter, L., Henseler, K. & Maier, B. Differential interaction forces govern bacterial sorting in early biofilms. eLife 4, e10811 (2015).
Smukalla, S. et al. FLO1 is a variable green beard gene that drives biofilm-like cooperation in budding yeast. Cell 135, 726–737 (2008).
Dawkins, R. The Selfish Gene (Oxford Univ. Press, 1989).
Maynard Smith, J. & Szathmary, E. The Major Transitions in Evolution (Oxford Univ. Press, 1995).
Tarnita, C. E., Taubes, C. H. & Nowak, M. A. Evolutionary construction by staying together and coming together. J. Theor. Biol. 320, 10–22 (2013).
Claessen, D., Rozen, D. E., Kuipers, O. P., Søgaard-Andersen, L. & van Wezel, G. P. Bacterial solutions to multicellularity: a tale of biofilms, filaments and fruiting bodies. Nat. Rev. Microbiol. 12, 115–124 (2014).
Ratcliff, W. C., Denison, R. F., Borrello, M. & Travisano, M. Experimental evolution of multicellularity. Proc. Natl Acad. Sci. USA 109, 1595–1600 (2012).
Koschwanez, J. H., Foster, K. R. & Murray, A. Improved use of a public good selects for the evolution of undifferentiated multicellularity. eLife 2, e00367 (2013).
Koschwanez, J. H., Foster, K. R. & Murray, A. W. Sucrose utilization in budding yeast as a model for the origin of undifferentiated multicellularity. PLoS Biol. 9, e1001122 (2011).
Justice, S. S., Hunstad, D. A., Cegelski, L. & Hultgren, S. J. Morphological plasticity as a bacterial survival strategy. Nat. Rev. Microbiol. 6, 162–168 (2008).
Persat, A., Stone, H. A. & Gitai, Z. The curved shape of Caulobacter crescentus enhances surface colonization in flow. Nat. Commun. 5, 3824 (2014).
Drescher, K. et al. Architectural transitions in Vibrio cholerae biofilms at single-cell resolution. Proc. Natl Acad. Sci. USA 113, E2066–E2072 (2016).
Teschler, J. K. et al. Living in the matrix: assembly and control of Vibrio cholerae biofilms. Nat. Rev. Microbiol. 13, 255–268 (2015).
Berk, V. et al. Molecular architecture and assembly principles of Vibrio cholerae biofilms. Science 337, 236–239 (2012).
Nadell, C. D., Drescher, K., Wingreen, N. S. & Bassler, B. L. Extracellular matrix structure governs invasion resistance in bacterial biofilms. ISME J. 9, 1700–1709 (2015).
Smith, D. R. et al. In situ proteolysis of the Vibrio cholerae matrix protein RbmA promotes biofilm recruitment. Proc. Natl Acad. Sci. USA 112, 10491–10496 (2015).
Costerton, J. W., Lewandowski, Z., Caldwell, D. E., Korber, D. R. & Lappinscott, H. M. Microbial biofilms. Annu. Rev. Microbiol. 49, 711–745 (1995).
Roberts, A. E., Kragh, K. N., Bjarnsholt, T. & Diggle, S. P. The limitations of in vitro experimentation in understanding biofilms and chronic infection. J. Mol. Biol. 427, 3646–3661 (2015).
Rusconi, R., Garren, M. & Stocker, R. Microfluidics expanding the frontiers of microbial ecology. Annu. Rev. Biophys. 43, 65–91 (2014).
Earle, K. A. et al. Quantitative imaging of gut microbiota spatial organization. Cell Host Microbe 18, 478–488 (2015).
Harrison, F., Muruli, A., Higgins, S. & Diggle, S. P. Development of an ex vivo porcine lung model for studying growth, virulence, and signaling of Pseudomonas aeruginosa. Infect. Immun. 82, 3312–3323 (2014).
Welch, J. L. M., Rossetti, B. J., Rieken, C. W., Dewhirst, F. E. & Borisy, G. G. Biogeography of a human oral microbiome at the micron scale. Proc. Natl Acad. Sci. USA 113, E791–E800 (2016).
Coyte, K. Z., Schluter, J. & Foster, K. R. The ecology of the microbiome: network, competition, and stability. Science 350, 663–666 (2015).
Hamilton, W. D. Altruism and related phenomena, mainly in social insects. Annu. Rev. Ecol. Evol. Syst. 3, 192–232 (1972).
Foster, K. R. & Wenseleers, T. A general model for the evolution of mutualisms. J. Evol. Biol. 19, 1283–1293 (2006).
Kreft, J. U., Picioreanu, C., Wimpenny, J. W. T. & van Loosdrecht, M. C. M. Individual-based modelling of biofilms. Microbiology 147, 2897–2912 (2001).
Xavier, J. B., Picioreanu, C. & van Loosdrecht, M. C. M. A framework for multidimensional modelling of activity and structure of multispecies biofilms. Environ. Microbiol. 7, 1085–1103 (2005).
Kreft, J. U. Biofilms promote altruism. Microbiology 150, 2751–2760 (2004). A landmark individual-based modelling study demonstrating how the spatial structure of cell lineages can promote the evolution of cooperation in biofilms.
Schluter, J. & Foster, K. R. The evolution of mutualism in gut microbiota via host epithelial selection. PLoS Biol. 10, e1001424 (2012).
Zhao, K. et al. Psl trails guide exploration and microcolony formation in Pseudomonas aeruginosa biofilms. Nature 497, 388–391 (2013).
Acknowledgements
The authors are grateful to A. Persat, A. Stacy, D. Cornforth, N. Oliveira, W. Kim, S. Diggle and two anonymous reviewers for providing comments on the manuscript. P. Singh and R. Hartmann provided invaluable help in the preparation of figure 4d. Work in the contributing laboratories was supported by European Research Council grant 242670 (K.R.F.), The Max Planck Society (K.D.), the Human Frontier Science Program (K.D.) and the Alexander von Humboldt Foundation (C.D.N.).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Glossary
- Microbiota
-
The community of microorganisms that live in association with a particular host organism (for example, the gut microbiota) or abiotic environment (for example, the soil microbiota).
- Social phenotypes
-
Phenotypes that exert an effect (either positive or negative) on the reproductive output of other individuals and which evolved, in part, because of this fitness effect that they exert.
- Type VI secretion system
-
(T6SS). A mechanism for killing neighbouring cells by the extension of a phage-tail-derived structure to putatively puncture adjacent cells and deliver toxic effectors.
- Dispersal
-
The process by which cells depart from a community, either individually or collectively. Dispersal can be active, in response to stresses such as nutrient limitation, or passive, owing to biofilm erosion by fluid flow.
- Genetic drift
-
A change in allele frequency in a population due to random sampling of organisms across generations (for example, due to stochasticity in reproductive success).
- Public goods
-
Substances that are secreted into the extracellular space that provide a benefit to other cells in the vicinity.
- Cheating mutants
-
Genotypes that gain a relative fitness advantage by receiving the benefits of an evolved cooperative trait of other genotypes, such as a public good, without contributing to the cooperative interaction themselves.
- Ecological productivity
-
The total biomass produced by a strain or species in a given environmental setting
- Antibiotics
-
Molecules that are produced by various microorganisms and act as toxins against other microorganisms; some antibiotics have been co-opted as pharmaceuticals for the treatment of microbial infections.
- Bacteriocins
-
Antibiotics that are produced by bacteria and specifically target other bacteria. Bacteriocins often occur as toxin–antitoxin pairs that are encoded on the same plasmid or in the same genomic neighbourhood.
- Contact-dependent inhibition
-
A mechanism of inhibiting the growth of neighbouring cells by the extension of a helical structure to contact target cells and deliver toxic effector molecules.
- Syntrophic relationships
-
Interactions in which one species benefits by using the product of another as a nutrient source; the producing species may in turn benefit from the removal of this product.
- Flocculation
-
Aggregation of yeast cells to form large multicellular groups that precipitate from liquid cultures and exhibit heightened stress tolerance.
- Greenbeard gene
-
A gene (or a set of closely linked genes) that is responsible for both an identifying phenotypic trait and a cooperative behaviour that targets that identifying trait, ensuring that the greenbeard gene bearer preferentially benefits other bearers of the greenbeard gene.
Rights and permissions
About this article
Cite this article
Nadell, C., Drescher, K. & Foster, K. Spatial structure, cooperation and competition in biofilms. Nat Rev Microbiol 14, 589–600 (2016). https://doi.org/10.1038/nrmicro.2016.84
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrmicro.2016.84
This article is cited by
-
Optogenetic spatial patterning of cooperation in yeast populations
Nature Communications (2024)
-
Advances in bioleaching of waste lithium batteries under metal ion stress
Bioresources and Bioprocessing (2023)
-
“Sharing the matrix” – a cooperative strategy for survival in Salmonella enterica serovar Typhimurium
BMC Microbiology (2023)
-
Starvation responses impact interaction dynamics of human gut bacteria Bacteroides thetaiotaomicron and Roseburia intestinalis
The ISME Journal (2023)
-
Dynamic social interactions and keystone species shape the diversity and stability of mixed-species biofilms – an example from dairy isolates
ISME Communications (2023)