Key Points
-
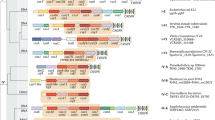
CRISPR–Cas systems form two major classes that differ in the organization of their effector modules. The effector modules of class 2 systems consist of a single large protein, which makes them the best candidates for genome-editing tools.
-
Computational methods of microbial genome screening were developed for the comprehensive identification of class 2 CRISPR–cas loci. Using these approaches, six new subtypes of the class 2 system were discovered, which brings the total for this class to three types and 10 subtypes.
-
Type II and type V CRISPR–Cas effectors are homologues of TnpB proteins, which are a poorly characterized family of nucleases that are encoded by bacterial and archaeal transposons. The different subtypes of these two types seem to have evolved independently, through the integration of TnpB-encoding transposons near CRISPR arrays.
-
Type VI effectors are large proteins that contain two RNase domains of the higher eukaryotes and prokaryotes nucleotide-binding domain (HEPN) superfamily and that have been shown to, or are predicted to, specifically target RNA.
-
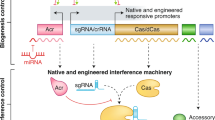
The diverse class 2 CRISPR–Cas systems that have been discovered provide opportunities for the construction of versatile genome-editing tools.
Abstract
Class 2 CRISPR–Cas systems are characterized by effector modules that consist of a single multidomain protein, such as Cas9 or Cpf1. We designed a computational pipeline for the discovery of novel class 2 variants and used it to identify six new CRISPR–Cas subtypes. The diverse properties of these new systems provide potential for the development of versatile tools for genome editing and regulation. In this Analysis article, we present a comprehensive census of class 2 types and class 2 subtypes in complete and draft bacterial and archaeal genomes, outline evolutionary scenarios for the independent origin of different class 2 CRISPR–Cas systems from mobile genetic elements, and propose an amended classification and nomenclature of CRISPR–Cas.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Makarova, K. S., Grishin, N. V., Shabalina, S. A., Wolf, Y. I. & Koonin, E. V. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol. Direct 1, 7 (2006).
Barrangou, R. et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712 (2007).
Barrangou, R. CRISPR–Cas systems and RNA-guided interference. Wiley Interdiscip. Rev. RNA 4, 267–278 (2013).
Marraffini, L. A. CRISPR–Cas immunity in prokaryotes. Nature 526, 55–61 (2015).
Mohanraju, P. et al. Diverse evolutionary roots and mechanistic variations of the CRISPR–Cas systems. Science 353, aad5147 (2016).
Van der Oost, J., Westra, E. R., Jackson, R. N. & Wiedenheft, B. Unravelling the structural and mechanistic basis of CRISPR–Cas systems. Nat. Rev. Microbiol. 12, 479–492 (2014).
Makarova, K. S., Aravind, L., Wolf, Y. I. & Koonin, E. V. Unification of Cas protein families and a simple scenario for the origin and evolution of CRISPR–Cas systems. Biol. Direct 6, 38 (2011).
Makarova, K. S., Wolf, Y. I. & Koonin, E. V. The basic building blocks and evolution of CRISPR–Cas systems. Biochem. Soc. Trans. 41, 1392–1400 (2013).
Takeuchi, N., Wolf, Y. I., Makarova, K. S. & Koonin, E. V. Nature and intensity of selection pressure on CRISPR-associated genes. J. Bacteriol. 194, 1216–1225 (2012).
Bondy-Denomy, J. & Davidson, A. R. To acquire or resist: the complex biological effects of CRISPR–Cas systems. Trends Microbiol. 22, 218–225 (2014).
Bondy-Denomy, J. et al. Multiple mechanisms for CRISPR–Cas inhibition by anti-CRISPR proteins. Nature 526, 136–139 (2015).
van Houte, S. et al. The diversity-generating benefits of a prokaryotic adaptive immune system. Nature 532, 385–388 (2016).
Makarova, K. S. et al. Evolution and classification of the CRISPR–Cas systems. Nat. Rev. Microbiol. 9, 467–477 (2011).
Makarova, K. S. & Koonin, E. V. Annotation and classification of CRISPR–Cas systems. Methods Mol. Biol. 1311, 47–75 (2015).
Makarova, K. S. et al. An updated evolutionary classification of CRISPR–Cas systems. Nat. Rev. Microbiol. 13, 722–736 (2015). This paper presents the latest classification of the CRISPR–Cas systems, prior to the application of the pipeline described here, along with computational approaches for the identification and quantitative comparison of CRISPR– cas loci.
Shmakov, S. et al. Discovery and functional characterization of diverse class 2 CRISPR–Cas systems. Mol. Cell 60, 385–397 (2015). This paper presents the first instalment of the computational pipeline that is described in this article, using Cas1 as the seed, and experimental validation of the activity of subtype V-B.
Chylinski, K., Makarova, K. S., Charpentier, E. & Koonin, E. V. Classification and evolution of type II CRISPR–Cas systems. Nucleic Acids Res. 42, 6091–6105 (2014).
Brouns, S. J. et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321, 960–964 (2008).
Jore, M. M. et al. Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nat. Struct. Mol. Biol. 18, 529–536 (2011).
Beloglazova, N. et al. CRISPR RNA binding and DNA target recognition by purified Cascade complexes from Escherichia coli. Nucleic Acids Res. 43, 530–543 (2015).
Jackson, R. N. et al. Structural biology. Crystal structure of the CRISPR RNA-guided surveillance complex from Escherichia coli. Science 345, 1473–1479 (2014).
Rouillon, C. et al. Structure of the CRISPR interference complex CSM reveals key similarities with cascade. Mol. Cell 52, 124–134 (2013).
Staals, R. H. et al. RNA targeting by the type III-A CRISPR–Cas Csm complex of Thermus thermophilus. Mol. Cell 56, 518–530 (2014).
Osawa, T., Inanaga, H., Sato, C. & Numata, T. Crystal structure of the CRISPR–Cas RNA silencing Cmr complex bound to a target analog. Mol. Cell 58, 418–430 (2015).
Taylor, D. W. et al. Structural biology. Structures of the CRISPR–Cmr complex reveal mode of RNA target positioning. Science 348, 581–585 (2015).
Jinek, M. et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012).
Cong, L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013).
Mali, P., Esvelt, K. M. & Church, G. M. Cas9 as a versatile tool for engineering biology. Nat. Methods 10, 957–963 (2013).
Gasiunas, G., Barrangou, R., Horvath, P. & Siksnys, V. Cas9–crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl Acad. Sci. USA 109, E2579–E2586 (2012).
Nishimasu, H. et al. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell 156, 935–949 (2014). This paper reports the first structure of Cas9, which provides insight into the interaction of class 2 effectors with crRNA and target DNA.
Nishimasu, H. et al. Crystal structure of Staphylococcus aureus Cas9. Cell 162, 1113–1126 (2015).
Sternberg, S. H., LaFrance, B., Kaplan, M. & Doudna, J. A. Conformational control of DNA target cleavage by CRISPR–Cas9. Nature 527, 110–113 (2015).
Sapranauskas, R. et al. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res. 39, 9275–9282 (2011).
Deltcheva, E. et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471, 602–607 (2011).
Chylinski, K., Le Rhun, A. & Charpentier, E. The tracrRNA and Cas9 families of type II CRISPR–Cas immunity systems. RNA Biol. 10, 726–737 (2013).
Briner, A. E. et al. Guide RNA functional modules direct Cas9 activity and orthogonality. Mol. Cell 56, 333–339 (2014).
Schunder, E., Rydzewski, K., Grunow, R. & Heuner, K. First indication for a functional CRISPR/Cas system in Francisella tularensis. Int. J. Med. Microbiol. 303, 51–60 (2013).
Zetsche, B. et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR–Cas system. Cell 163, 759–771 (2015). This work demonstrates the interference activity of Cpf1 and shows that Cpf1 is a single RNA-guided endonuclease that does not require tracrRNA.
Dong, D. et al. The crystal structure of Cpf1 in complex with CRISPR RNA. Nature 532, 522–526 (2016).
Yamano, T. et al. Crystal structure of Cpf1 in complex with guide RNA and target DNA. Cell 165, 949–962 (2016). Together with reference 39, this paper presents the structure of Cpf1 in complex with crRNA and target DNA, demonstrating that, despite similar overall shapes, the domain architectures of Cpf1 and Cas9 differ substantially.
Fonfara, I., Richter, H., Bratovic, M., Le Rhun, A. & Charpentier, E. The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature 532, 517–521 (2016). This work demonstrates that Cpf1 is responsible not only for interference but also for pre-crRNA processing.
Abudayyeh, O. O. et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 353, aaf5573 (2016). This paper describes the first CRISPR–Cas system that exclusively cleaves RNA, and demonstrates the switch from specific to non-specific RNA cleavage following target recognition.
Pasternak, C. et al. ISDra2 transposition in Deinococcus radiodurans is downregulated by TnpB. Mol. Microbiol. 88, 443–455 (2013).
Bao, W. & Jurka, J. Homologues of bacterial TnpB_IS605 are widespread in diverse eukaryotic transposable elements. Mob. DNA 4, 12 (2013).
Kapitonov, V. V., Makarova, K. S. & Koonin, E. V. ISC, a novel group of bacterial and archaeal DNA transposons that encode Cas9 homologs. J. Bacteriol. 198, 797–807 (2015). In this work, the direct evolutionary ancestors of Cas9 are identified.
Gomes-Filho, J. V. et al. Sense overlapping transcripts in IS1341-type transposase genes are functional non-coding RNAs in archaea. RNA Biol. 12, 490–500 (2015). This work demonstrates that TnpB proteins bind to RNA, which is compatible with their role as ancestors of class 2 CRISPR–Cas effectors.
Westra, E. R., Buckling, A. & Fineran, P. C. CRISPR–Cas systems: beyond adaptive immunity. Nat. Rev. Microbiol. 12, 317–326 (2014).
Anantharaman, V., Makarova, K. S., Burroughs, A. M., Koonin, E. V. & Aravind, L. Comprehensive analysis of the HEPN superfamily: identification of novel roles in intra-genomic conflicts, defense, pathogenesis and RNA processing. Biol. Direct 8, 15 (2013).
Makarova, K. S., Anantharaman, V., Grishin, N. V., Koonin, E. V. & Aravind, L. CARF and WYL domains: ligand-binding regulators of prokaryotic defense systems. Front. Genet. 5, 102 (2014).
Makarova, K. S., Anantharaman, V., Aravind, L. & Koonin, E. V. Live virus-free or die: coupling of antivirus immunity and programmed suicide or dormancy in prokaryotes. Biol. Direct 7, 40 (2012).
Iranzo, J., Lobkovsky, A. E., Wolf, Y. I. & Koonin, E. V. Immunity, suicide or both? Ecological determinants for the combined evolution of anti-pathogen defense systems. BMC Evol. Biol. 15, 43 (2015).
East-Seletsky, A. et al. Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature 538, 270–273 (2016). This paper describes experiments that show that, similar to Cpf1, C2c2, the subtype VI-A effector, catalyses pre-crRNA processing.
Koonin, E. V., Dolja, V. V. & Krupovic, M. Origins and evolution of viruses of eukaryotes: the ultimate modularity. Virology 479–480, 2–25 (2015).
Sheppard, N. F., Glover, C. V., Terns, R. M. & Terns, M. P. The CRISPR-associated Csx1 protein of Pyrococcus furiosus is an adenosine-specific endoribonuclease. RNA 22, 216–224 (2016).
Niewoehner, O. & Jinek, M. Structural basis for the endoribonuclease activity of the type III-A CRISPR-associated protein Csm6. RNA 22, 318–329 (2016).
Curtis, T. P., Sloan, W. T. & Scannell, J. W. Estimating prokaryotic diversity and its limits. Proc. Natl Acad. Sci. USA 99, 10494–10499 (2002).
Curtis, T. P. et al. What is the extent of prokaryotic diversity? Philos. Trans. R. Soc. Lond. B Biol. Sci. 361, 2023–2037 (2006).
Fraser, C., Alm, E. J., Polz, M. F., Spratt, B. G. & Hanage, W. P. The bacterial species challenge: making sense of genetic and ecological diversity. Science 323, 741–746 (2009).
Quince, C., Curtis, T. P. & Sloan, W. T. The rational exploration of microbial diversity. ISME J. 2, 997–1006 (2008).
Chavez, A. et al. Comparison of Cas9 activators in multiple species. Nat. Methods 13, 563–567 (2016).
Thakore, P. I., Black, J. B., Hilton, I. B. & Gersbach, C. A. Editing the epigenome: technologies for programmable transcription and epigenetic modulation. Nat. Methods 13, 127–137 (2016).
Chen, B. et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell 155, 1479–1491 (2013).
Knight, S. C. et al. Dynamics of CRISPR–Cas9 genome interrogation in living cells. Science 350, 823–826 (2015).
Nelles, D. A. et al. Programmable RNA tracking in live cells with CRISPR/Cas9. Cell 165, 488–496 (2016).
Hsu, P. D., Lander, E. S. & Zhang, F. Development and applications of CRISPR–Cas9 for genome engineering. Cell 157, 1262–1278 (2014).
Kleinstiver, B. P. et al. Genome-wide specificities of CRISPR–Cas Cpf1 nucleases in human cells. Nat. Biotechnol. 34, 869–874 (2016).
Kim, Y. et al. Generation of knockout mice by Cpf1-mediated gene targeting. Nat. Biotechnol. 34, 808–810 (2016).
Hur, J. K. et al. Targeted mutagenesis in mice by electroporation of Cpf1 ribonucleoproteins. Nat. Biotechnol. 34, 807–808 (2016).
Kim, D. et al. Genome-wide analysis reveals specificities of Cpf1 endonucleases in human cells. Nat. Biotechnol. 34, 863–868 (2016).
Li, S. Y., Zhao, G. P. & Wang, J. C-Brick: a new standard for assembly of biological parts using Cpf1. ACS Synth. Biol. http://dx.doi.org/10.1021/acssynbio.6b00114 (2016).
O'Connell, M. R. et al. Programmable RNA recognition and cleavage by CRISPR/Cas9. Nature 516, 263–266 (2014).
Edgar, R. C. PILER-CR: fast and accurate identification of CRISPR repeats. BMC Bioinformatics 8, 18 (2007).
Grissa, I., Vergnaud, G. & Pourcel, C. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 35, W52–W57 (2007).
Almendros, C., Guzman, N. M., Garcia-Martinez, J. & Mojica, F. J. Anti-cas spacers in orphan CRISPR4 arrays prevent uptake of active CRISPR–Cas I-F systems. Nat. Microbiol. 1, 16081 (2016).
Makarova, K. S., Wolf, Y. I. & Koonin, E. V. Comparative genomics of defense systems in archaea and bacteria. Nucleic Acids Res. 41, 4360–4377 (2013).
Liu, L. et al. C2c1–sgRNA complex structure reveals RNA-guided DNA cleavage mechanism. Mol. Cell http://dx.doi.org/10.1016/j.molcel.2016.11.040 (2016).
Yang, H., Gao, P., Rajashankar, K. R. & Patel, D. J. PAM-dependent target DNA recognition and cleavage by C2c1 CRISPR–Cas endonuclease. Cell 167, 1814–1828.e12 (2016).
Liu, L. et al. Two distant catalytic sites are responsible for C2c2 RNase activities. Cell 168, 121–134.e12. (2017).
Smargon, A. A. et al. Cas13b is a type VI-B CRISPR-associated RNA-guided RNase differentially regulated by accessory proteins Csx27 and Csx28. Mol. Cell http://dx.doi.org/10.1016/j.molcel.2016.12.023 (2017).
Burstein, D. et al. New CRISPR–Cas systems from uncultivated microbes. Nature http://dx.doi.org/10.1038/nature21059 (2016).
Acknowledgements
S.S., K.S.M., Y.I.W. and E.V.K. are supported by intramural funds of the US Department of Health and Human Services (to the US National Library of Medicine). K.S. is supported by a US National Institutes of Health (NIH) grant (GM10407), a Russian Science Foundation grant (14-14-00988), and by the Skolkovo Institute of Science and Technology (Skoltech). F.Z. is supported by the NIH through the US National Institute of Mental Health (NIMH; grants 5DP1-MH100706 and 1R01-MH110049); the US National Science Foundation (NSF); the New York Stem Cell Foundation; the Allen Distinguished Investigator Program; through The Paul G. Allen Frontiers Group; the Simons and Vallee Foundations; the Howard Hughes Medical Institute; the Skoltech–MIT Next Generation Program; James and Patricia Poitras and the Poitras Center for Affective Disorders; R. Metcalfe; and D. Cheng. F.Z. is a New York Stem Cell Foundation–Robertson Investigator.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
F.Z. is a co-founder of, and scientific adviser for, Editas Medicine and is a scientific adviser for Horizon Discovery.
Related links
Supplementary information
Supplementary information S1 (box)
Supplementary Methods description. (PDF 158 kb)
Supplementary information S2 (box)
Supplementary Excel Files (PDF 100 kb)
Supplementary information S3 (figure)
Multiple alignment of representatives from five V-U families. (PDF 1628 kb)
Supplementary information S4 (figure)
Strength of purifying selection for Class 2 effector protein families. (PDF 90 kb)
Supplementary information S5 (figure)
UPGMA dendrogram of protein family profile similarity (PDF 136 kb)
Supplementary information S6 (figure)
Membrane proteins associated with Cas13b genes (PDF 153 kb)
Glossary
- CRISPR–Cas
-
An adaptive immune system in archaea and bacteria that functions by inserting fragments of foreign genomes into CRISPR arrays and using the transcripts of the resulting spacers as guide RNAs to detect and inactivate the cognate genetic elements.
- Adaptation
-
The first phase of the CRISPR immune response, during which a piece of foreign DNA is inserted into a CRISPR array to become a spacer that is subsequently used as the template to produce the CRISPR RNA (crRNA).
- CRISPR RNAs
-
(crRNAs). Small RNA molecules that consist of the RNA complement of a spacer and parts of the two adjacent repeats. crRNAs are produced by processing of the transcript of the entire CRISPR array (pre-crRNA); processing is mediated either by Cas proteins only (class 1, type V-A and type VI-A systems) or by an external RNase, such as bacterial RNase III, in conjunction with Cas proteins.
- Interference
-
The final phase of the CRISPR immune response, during which the target DNA (or less commonly, RNA) is recognized by a CRISPR effector through the bound CRISPR RNA (crRNA) and cleaved by the effector nuclease or nucleases.
- Effector
-
A complex of Cas proteins (in class 1 systems), or a single, large protein (in class 2 systems), that is involved in target recognition and inactivation, and, in most cases, in the processing of pre-CRISPR RNA (pre-crRNA).
- Class 2 CRISPR–Cas systems
-
One of the two major divisions of CRISPR–Cas that is characterized by effector modules that consist of a single, large protein with endonuclease activity.
- Trans-acting CRISPR RNA
-
(tracrRNA). An accessory RNA molecule that is partially complementary to CRISPR and is involved in pre-crRNA processing in type II and type V-B CRISPR–Cas interference.
- Higher eukaryotes and prokaryotes nucleotide-binding domains
-
(HEPN domains). An early name that was given when the functions of the domains were not well characterized. An expansive superfamily of domains with RNase activity that are involved in various defence functions, in particular, type VI and some class 1 CRISPR–Cas interference.
- TnpB proteins
-
A poorly characterized superfamily of transposon-encoded proteins that contain RuvC-like nuclease domains. TnpB proteins are the apparent evolutionary ancestors of type II and type V CRISPR–Cas effectors.
Rights and permissions
About this article
Cite this article
Shmakov, S., Smargon, A., Scott, D. et al. Diversity and evolution of class 2 CRISPR–Cas systems. Nat Rev Microbiol 15, 169–182 (2017). https://doi.org/10.1038/nrmicro.2016.184
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrmicro.2016.184
This article is cited by
-
Comprehensive review of CRISPR-based gene editing: mechanisms, challenges, and applications in cancer therapy
Molecular Cancer (2024)
-
A novel detection method based on MIRA-CRISPR/Cas13a-LFD targeting the repeated DNA sequence of Trichomonas vaginalis
Parasites & Vectors (2024)
-
Precise genome-editing in human diseases: mechanisms, strategies and applications
Signal Transduction and Targeted Therapy (2024)
-
Efficient engineering of human and mouse primary cells using peptide-assisted genome editing
Nature Biotechnology (2024)
-
CRISPR–Cas13d in plant biology: an insight
Plant Biotechnology Reports (2024)