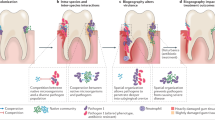
Key Points
-
The human microbiome and invading pathogens colonize the host as spatially organized polymicrobial (multi-species or multi-strain) communities, exhibiting spatial patterning or 'biogeography' both at the macroscale (across the human body) and microscale levels (within individual infection sites).
-
Host landmarks that drive biogeography include receptors for microbial attachment, physicochemical cues of pH and oxygen, host-derived nutrients, and the immune response.
-
Specific spatial organizations that result from host–microorganism interactions include localized attachment to host surfaces, metabolically optimal positioning along physicochemical or nutritional gradients, and the enforced segregation of the microbiome from the host by the immune system.
-
Polymicrobial interactions also drive spatial organization in multispecies biofilms, generally resulting in either mixed or segregated spatial patterns, depending on the extent of cooperation or competition between community members.
-
Specific polymicrobial interactions that drive microbiogeography include intercellular attachment (within and between species), biofilm remodelling (by making or breaking down the supporting matrix), and the export of diffusible molecules that promote or suppress the growth of neighbouring cells.
-
Spatial organization and the determining community interactions can, in many cases, alter the progression of infection, in what is known as the 'biogeography of infection.'
Abstract
Microbial communities are spatially organized in both the environment and the human body. Although patterns exhibited by these communities are described by microbial biogeography, this discipline has previously only considered large-scale, global patterns. By contrast, the fine-scale positioning of a pathogen within an infection site can greatly alter its virulence potential. In this Review, we highlight the importance of considering spatial positioning in the study of polymicrobial infections and discuss targeting biogeography as a therapeutic strategy.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Hall-Stoodley, L., Costerton, J. W. & Stoodley, P. Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2, 95–108 (2004).
Kaufmann, S. H. & Schaible, U. E. 100th anniversary of Robert Koch's Nobel Prize for the discovery of the tubercle bacillus. Trends Microbiol. 13, 469–475 (2005).
Smith, H. The role of microbial interactions in infectious disease. Phil. Trans. R. Soc. Lond. B. Biol. Sci. 297, 551–561 (1982).
Hajishengallis, G. et al. Low-abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and complement. Cell Host Microbe 10, 497–506 (2011).
Korgaonkar, A., Trivedi, U., Rumbaugh, K. P. & Whiteley, M. Community surveillance enhances Pseudomonas aeruginosa virulence during polymicrobial infection. Proc. Natl Acad. Sci. USA 110, 1059–1064 (2013).
Simon-Soro, A. & Mira, A. Solving the etiology of dental caries. Trends Microbiol. 23, 76–82 (2015).
Murray, J. L., Connell, J. L., Stacy, A., Turner, K. H. & Whiteley, M. Mechanisms of synergy in polymicrobial infections. J. Microbiol. 52, 188–199 (2014).
Sibley, C. D. et al. Discerning the complexity of community interactions using a Drosophila model of polymicrobial infections. PLoS Pathog. 4, e1000184 (2008). This study defined the major categories of polymicrobial virulence, including additive and synergistic, by screening P. aeruginosa with 40 respiratory tract isolates in a Drosophila co-infection model.
Onderdonk, A. B., Bartlett, J. G., Louie, T., Sullivan-Seigler, N. & Gorbach, S. L. Microbial synergy in experimental intra-abdominal abscess. Infect. Immun. 13, 22–26 (1976).
Trivedi, U. et al. Prevalence of multiple antibiotic resistant infections in diabetic versus nondiabetic wounds. J. Pathog. 2014, 173053 (2014).
Hogan, D. A. & Kolter, R. Pseudomonas–Candida interactions: an ecological role for virulence factors. Science 296, 2229–2232 (2002).
Griffiths, E. C., Pedersen, A. B., Fenton, A. & Petchey, O. L. The nature and consequences of coinfection in humans. J. Infect. 63, 200–206 (2011).
Whitmore, T. C. (ed) Wallace's Line and Plate Tectonics (Oxford: Clarendon Press; Oxford University Press; 1981).
Vellend, M. Conceptual synthesis in community ecology. Q. Rev. Biol. 85, 183–206 (2010).
Nemergut, D. R. et al. Patterns and processes of microbial community assembly. Microbiol. Mol. Biol. Rev. 77, 342–356 (2013).
Hanson, C. A., Fuhrman, J. A., Horner-Devine, M. C. & Martiny, J. B. Beyond biogeographic patterns: processes shaping the microbial landscape. Nat. Rev. Microbiol. 10, 497–506 (2012).
Costello, E. K. et al. Bacterial community variation in human body habitats across space and time. Science 326, 1694–1697 (2009).
Stearns, J. C. et al. Bacterial biogeography of the human digestive tract. Sci. Rep. 1, 170 (2011).
Oh, J. et al. Biogeography and individuality shape function in the human skin metagenome. Nature 514, 59–64 (2014).
Vos, M., Wolf, A. B., Jennings, S. J. & Kowalchuk, G. A. Micro-scale determinants of bacterial diversity in soil. FEMS Microbiol. Rev. 37, 936–954 (2013).
Kerr, B., Riley, M. A., Feldman, M. W. & Bohannan, B. J. Local dispersal promotes biodiversity in a real-life game of rock-paper-scissors. Nature 418, 171–174 (2002).
Kim, H. J., Boedicker, J. Q., Choi, J. W. & Ismagilov, R. F. Defined spatial structure stabilizes a synthetic multispecies bacterial community. Proc. Natl Acad. Sci. USA 105, 18188–18193 (2008).
Drescher, K., Nadell, C. D., Stone, H. A., Wingreen, N. S. & Bassler, B. L. Solutions to the public goods dilemma in bacterial biofilms. Curr. Biol. 24, 50–55 (2014).
Zhou, L., Slamti, L., Nielsen-LeRoux, C., Lereclus, D. & Raymond, B. The social biology of quorum sensing in a naturalistic host pathogen system. Curr. Biol. 24, 2417–2422 (2014).
Harding, J. L. & Reynolds, M. M. Combating medical device fouling. Trends Biotechnol. 32, 140–146 (2014).
Palmer, R. J. Jr., Gordon, S. M., Cisar, J. O. & Kolenbrander, P. E. Coaggregation-mediated interactions of streptococci and actinomyces detected in initial human dental plaque. J. Bacteriol. 185, 3400–3409 (2003).
Gunning, A. P. et al. Mining the “glycocode” —exploring the spatial distribution of glycans in gastrointestinal mucin using force spectroscopy. FASEB J. 27, 2342–2354 (2013).
Melican, K. et al. Uropathogenic Escherichia coli P and type 1 fimbriae act in synergy in a living host to facilitate renal colonization leading to nephron obstruction. PLoS Pathog. 7, e1001298 (2011).
Bjarnsholt, T. et al. The in vivo biofilm. Trends Microbiol. 21, 466–474 (2013).
Schaber, J. A. et al. Pseudomonas aeruginosa forms biofilms in acute infection independent of cell-to-cell signaling. Infect. Immun. 75, 3715–3721 (2007).
Dalton, T. et al. An in vivo polymicrobial biofilm wound infection model to study interspecies interactions. PLoS ONE 6, e27317 (2011).
Stacy, A. et al. Bacterial fight-and-flight responses enhance virulence in a polymicrobial infection. Proc. Natl Acad. Sci. USA 111, 7819–7824 (2014).
Singh, P. K. et al. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407, 762–764 (2000).
Nguyen, C. T. et al. Noninvasive in vivo optical detection of biofilm in the human middle ear. Proc. Natl Acad. Sci. USA 109, 9529–9534 (2012).
Monds, R. D. & O'Toole, G. A. The developmental model of microbial biofilms: ten years of a paradigm up for review. Trends Microbiol. 17, 73–87 (2009).
Tran, C. S. et al. Translocon is required for biofilm formation at the epithelial barrier. PLoS Pathog. 10, e1004479 (2014). This study identified a determinant for P. aeruginosa biofilm formation in vivo that is dispensable in vitro , illustrating that microorganisms should be studied in natural and infectious contexts to gain better insight into the mechanisms of spatial organization.
Travier, L. et al. ActA promotes Listeria monocytogenes aggregation, intestinal colonization and carriage. PLoS Pathog. 9, e1003131 (2013).
Blanchette-Cain, K. et al. Streptococcus pneumoniae biofilm formation is strain dependent, multifactorial, and associated with reduced invasiveness and immunoreactivity during colonization. mBio 4, e00745-13 (2013).
Davies, D. G. et al. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280, 295–298 (1998).
Millet, Y. A. et al. Insights into Vibrio cholerae intestinal colonization from monitoring fluorescently labeled bacteria. PLoS Pathog. 10, e1004405 (2014).
Nadell, C. D., Foster, K. R. & Xavier, J. B. Emergence of spatial structure in cell groups and the evolution of cooperation. PLoS Comput. Biol. 6, e1000716 (2010).
Jemielita, M. et al. Spatial and temporal features of the growth of a bacterial species colonizing the zebrafish gut. mBio 5, e01751-14 (2014).
Monier, J. M. & Lindow, S. E. Differential survival of solitary and aggregated bacterial cells promotes aggregate formation on leaf surfaces. Proc. Natl Acad. Sci. USA 100, 15977–15982 (2003).
Connell, J. L. et al. Probing prokaryotic social behaviors with bacterial “lobster traps”. mBio 1, e00202-10 (2010).
Guggenberger, C., Wolz, C., Morrissey, J. A. & Heesemann, J. Two distinct coagulase-dependent barriers protect Staphylococcus aureus from neutrophils in a three dimensional in vitro infection model. PLoS Pathog. 8, e1002434 (2012).
Kragh, K. N. et al.: Polymorphonuclear leukocytes restrict growth of Pseudomonas aeruginosa in the lungs of cystic fibrosis patients. Infect. Immun. 82, 4477–4486 (2014). This study illustrated how microbiogeography (the local suppression of microbial growth by neutrophils) can determine macrobiogeography (in which aggregates of P. aeruginosa preferentially colonize the respiratory tract).
Davis, K. M., Mohammadi, S. & Isberg, R. R. Community behavior and spatial regulation within a bacterial microcolony in deep tissue sites serves to protect against host attack. Cell Host Microbe 17, 21–31 (2015). This study examined how microcolonies of Yersinia pseudotuberculosis in the spleen repel attacking neutrophils, illustrating how bacterial aggregates can adeptly resist the immune system.
Faruque, S. M. et al. Transmissibility of cholera: in vivo-formed biofilms and their relationship to infectivity and persistence in the environment. Proc. Natl Acad. Sci. USA 103, 6350–6355 (2006).
Singh, P. K., Parsek, M. R., Greenberg, E. P. & Welsh, M. J. A component of innate immunity prevents bacterial biofilm development. Nature 417, 552–555 (2002).
Caldara, M. et al. Mucin biopolymers prevent bacterial aggregation by retaining cells in the free-swimming state. Curr. Biol. 22, 2325–2330 (2012).
Alhede, M. et al. Pseudomonas aeruginosa recognizes and responds aggressively to the presence of polymorphonuclear leukocytes. Microbiology 155, 3500–3508 (2009).
Schreiber, S. et al. The spatial orientation of Helicobacter pylori in the gastric mucus. Proc. Natl Acad. Sci. USA 101, 5024–5029 (2004).
Falush, D. et al. Recombination and mutation during long-term gastric colonization by Helicobacter pylori: estimates of clock rates, recombination size, and minimal age. Proc. Natl Acad. Sci. USA 98, 15056–15061 (2001).
Yasuda, K. et al. Biogeography of the intestinal mucosal and lumenal microbiome in the rhesus macaque. Cell Host Microbe 17, 385–391 (2015).
Marteyn, B. et al. Modulation of Shigella virulence in response to available oxygen in vivo. Nature 465, 355–358 (2010).
Wessel, A. K. et al. Oxygen limitation within a bacterial aggregate. mBio 5, e00992 (2014).
Bradshaw, D. J., Marsh, P. D., Watson, G. K. & Allison, C. Role of Fusobacterium nucleatum and coaggregation in anaerobe survival in planktonic and biofilm oral microbial communities during aeration. Infect. Immun. 66, 4729–4732 (1998).
Bowler, P. G., Duerden, B. I. & Armstrong, D. G. Wound microbiology and associated approaches to wound management. Clin. Microbiol. Rev. 14, 244–269 (2001).
Turner, K. H., Everett, J., Trivedi, U., Rumbaugh, K. P. & Whiteley, M. Requirements for Pseudomonas aeruginosa acute burn and chronic surgical wound infection. PLoS Genet. 10, e1004518 (2014).
Berry, D. et al. Host-compound foraging by intestinal microbiota revealed by single-cell stable isotope probing. Proc. Natl Acad. Sci. USA 110, 4720–4725 (2013).
Schluter, J. & Foster, K. R. The evolution of mutualism in gut microbiota via host epithelial selection. PLoS Biol. 10, e1001424 (2012).
Nielsen, A. T. et al. A bistable switch and anatomical site control Vibrio cholerae virulence gene expression in the intestine. PLoS Pathog. 6, e1001102 (2010).
Kamada, N. et al. Regulated virulence controls the ability of a pathogen to compete with the gut microbiota. Science 336, 1325–1329 (2012).
Rivera-Chavez, F. et al. Salmonella uses energy taxis to benefit from intestinal inflammation. PLoS Pathog. 9, e1003267 (2013).
Thiennimitr, P. et al. Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc. Natl Acad. Sci. USA 108, 17480–17485 (2011).
Winter, S. E. et al. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467, 426–429 (2010).
Leveau, J. H. & Lindow, S. E. Appetite of an epiphyte: quantitative monitoring of bacterial sugar consumption in the phyllosphere. Proc. Natl Acad. Sci. USA 98, 3446–3453 (2001).
Lee, S. M. et al. Bacterial colonization factors control specificity and stability of the gut microbiota. Nature 501, 426–429 (2013). This study showed how microbiogeography (targeting of B. fragilis to the intestinal crypts) can contribute to stable colonization of the host.
Vaishnava, S. et al. The antibacterial lectin RegIIIγ promotes the spatial segregation of microbiota and host in the intestine. Science 334, 255–258 (2011).
Johansson, M. E. et al. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc. Natl Acad. Sci. USA 105, 15064–15069 (2008).
Johansson, M. E. & Hansson, G. C. Microbiology. keeping bacteria at a distance. Science 334, 182–183 (2011).
Lidell, M. E., Moncada, D. M., Chadee, K. & Hansson, G. C. Entamoeba histolytica cysteine proteases cleave the MUC2 mucin in its C-terminal domain and dissolve the protective colonic mucus gel. Proc. Natl Acad. Sci. USA 103, 9298–9303 (2006).
Chau, T. A. et al. Toll-like receptor 2 ligands on the staphylococcal cell wall downregulate superantigen-induced T cell activation and prevent toxic shock syndrome. Nat. Med. 15, 641–648 (2009).
Round, J. L. et al. The Toll-Like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science 332, 974–977 (2011).
Bergstrom, K. S. et al. Muc2 protects against lethal infectious colitis by disassociating pathogenic and commensal bacteria from the colonic mucosa. PLoS Pathog. 6, e1000902 (2010).
Velcich, A. et al. Colorectal cancer in mice genetically deficient in the mucin Muc2. Science 295, 1726–1729 (2002).
Dejea, C. M. et al. Microbiota organization is a distinct feature of proximal colorectal cancers. Proc. Natl Acad. Sci. USA 111, 18321–18326 (2014).
Momeni, B., Brileya, K. A., Fields, M. W. & Shou, W. Strong inter-population cooperation leads to partner intermixing in microbial communities. eLIFE 2, e00230 (2013).
Chalmers, N. I., Palmer, R. J. Jr., Cisar, J. O. & Kolenbrander, P. E. Characterization of a Streptococcus sp.-Veillonella sp. community micromanipulated from dental plaque. J. Bacteriol. 190, 8145–8154 (2008).
Estrela, S. & Brown, S. P. Metabolic and demographic feedbacks shape the emergent spatial structure and function of microbial communities. PLoS Comput. Biol. 9, e1003398 (2013). This study used individual-based modelling to show that the strength of metabolic dependency determines the level of mixing in microbial communities.
Zijnge, V. et al. Oral biofilm architecture on natural teeth. PLoS ONE 5, e9321 (2010).
Kolenbrander, P. E., Palmer, R. J. Jr., Periasamy, S. & Jakubovics, N. S. Oral multispecies biofilm development and the key role of cell-cell distance. Nat. Rev. Microbiol. 8, 471–480 (2010).
Valm, A. M. et al. Systems-level analysis of microbial community organization through combinatorial labeling and spectral imaging. Proc. Natl Acad. Sci. USA 108, 4152–4157 (2011). This study presents a 'nearest neighbour' network, in which frequencies of intercellular associations are described, for 15 taxonomic groups in human dental plaque.
Egland, P. G., Palmer, R. J. Jr., Kolenbrander, P. E. Interspecies communication in Streptococcus gordonii-Veillonella atypica biofilms: signaling in flow conditions requires juxtaposition. Proc. Natl Acad. Sci. USA 101, 16917–16922 (2004).
Jakubovics, N. S., Gill, S. R., Iobst, S. E., Vickerman, M. M. & Kolenbrander, P. E. Regulation of gene expression in a mixed-genus community: stabilized arginine biosynthesis in Streptococcus gordonii by coaggregation with Actinomyces naeslundii. J. Bacteriol. 190, 3646–3657 (2008).
He, X. et al. Cultivation of a human-associated TM7 phylotype reveals a reduced genome and epibiotic parasitic lifestyle. Proc. Natl Acad. Sci. USA 112, 244–249 (2015).
Kim, W., Racimo, F., Schluter, J., Levy, S. B. & Foster, K. R. Importance of positioning for microbial evolution. Proc. Natl Acad. Sci. USA 111, E1639–E1647 (2014). This study showed how inter-strain competition can select for novel positioning strategies in biofilms, such as hyper-secretion to push above neighbours.
Markussen, T. et al. Environmental heterogeneity drives within-host diversification and evolution of Pseudomonas aeruginosa. mBio 5, e01592-14 (2014).
Yang, L. et al. Pattern differentiation in co-culture biofilms formed by Staphylococcus aureus and Pseudomonas aeruginosa. FEMS Immunol. Med. Microbiol. 62, 339–347 (2011).
Chew, S. C. et al. Dynamic remodeling of microbial biofilms by functionally distinct exopolysaccharides. mBio 5, e01536-14 (2014).
Huse, H. K. et al. Pseudomonas aeruginosa enhances production of a non-alginate exopolysaccharide during long-term colonization of the cystic fibrosis lung. PLoS ONE 8, e82621 (2013).
Eberl, L. & Tummler, B. Pseudomonas aeruginosa and Burkholderia cepacia in cystic fibrosis: genome evolution, interactions and adaptation. Int. J. Med. Microbiol. 294, 123–131 (2004).
Bragonzi, A. et al. Modelling co-infection of the cystic fibrosis lung by Pseudomonas aeruginosa and Burkholderia cenocepacia reveals influences on biofilm formation and host response. PLoS ONE 7, e52330 (2012).
Ryan, R. P. et al. Interspecies signalling via the Stenotrophomonas maltophilia diffusible signal factor influences biofilm formation and polymyxin tolerance in Pseudomonas aeruginosa. Mol. Microbiol. 68, 75–86 (2008).
Twomey, K. B. et al. Bacterial cis-2-unsaturated fatty acids found in the cystic fibrosis airway modulate virulence and persistence of Pseudomonas aeruginosa. ISME J. 6, 939–950 (2012).
Armbruster, C. E. et al. Indirect pathogenicity of Haemophilus influenzae and Moraxella catarrhalis in polymicrobial otitis media occurs via interspecies quorum signaling. mBio 1, e00102–10 (2014).
Park, S. et al. Motion to form a quorum. Science 301, 188 (2003).
Davies, D. G. & Marques, C. N. A fatty acid messenger is responsible for inducing dispersion in microbial biofilms. J. Bacteriol. 191, 1393–1403 (2009).
Vega, N. M., Allison, K. R., Samuels, A. N., Klempner, M. S. & Collins, J. J. Salmonella typhimurium intercepts Escherichia coli signaling to enhance antibiotic tolerance. Proc. Natl Acad. Sci. USA 110, 14420–14425 (2013).
Connell, J. L., Kim, J., Shear, J. B., Bard, A. J. & Whiteley, M. Real-time monitoring of quorum sensing in 3D-printed bacterial aggregates using scanning electrochemical microscopy. Proc. Natl Acad. Sci. USA 111, 18255–18260 (2014).
Liu, X. et al. Real-time mapping of a hydrogen peroxide concentration profile across a polymicrobial bacterial biofilm using scanning electrochemical microscopy. Proc. Natl Acad. Sci. USA 108, 2668–2673 (2011).
Xiao, J. et al. The exopolysaccharide matrix modulates the interaction between 3D architecture and virulence of a mixed-species oral biofilm. PLoS Pathog. 8, e1002623 (2012).
von Ohle, C. et al. Real-time microsensor measurement of local metabolic activities in ex vivo dental biofilms exposed to sucrose and treated with chlorhexidine. Appl. Environ. Microbiol. 76, 2326–2334 (2010).
Fazli, M. et al. Nonrandom distribution of Pseudomonas aeruginosa and Staphylococcus aureus in chronic wounds. J. Clin. Microbiol. 47, 4084–4089 (2009).
Hutchison, J. B. et al. Single-cell control of initial spatial structure in biofilm development using laser trapping. Langmuir 30, 4522–4530 (2014).
DeLeon, S. et al. Synergistic interactions of Pseudomonas aeruginosa and Staphylococcus aureus in an in vitro wound model. Infect. Immun. 82, 4718–4728 (2014).
Kirkup, B. C. & Riley, M. A. Antibiotic-mediated antagonism leads to a bacterial game of rock-paper-scissors in vivo. Nature 428, 412–414 (2004).
Cowan, S. E., Gilbert, E., Liepmann, D. & Keasling, J. D. Commensal interactions in a dual-species biofilm exposed to mixed organic compounds. Appl. Environ. Microbiol. 66, 4481–4485 (2000).
Nielsen, A. T., Tolker-Nielsen, T., Barken, K. B. & Molin, S. Role of commensal relationships on the spatial structure of a surface-attached microbial consortium. Environ. Microbiol. 2, 59–68 (2000).
Leriche, V., Briandet, R. & Carpentier, B. Ecology of mixed biofilms subjected daily to a chlorinated alkaline solution: spatial distribution of bacterial species suggests a protective effect of one species to another. Environ. Microbiol. 5, 64–71 (2003).
Connell, J. L., Ritschdorff, E. T., Whiteley, M. & Shear, J. B. 3D printing of microscopic bacterial communities. Proc. Natl Acad. Sci. USA 110, 18380–18385 (2013). This study illustrated how the spatial organization of multispecies communities can determine important virulence properties, such as antibiotic resistance.
Schillinger, C. et al. Co-localized or randomly distributed? Pair cross correlation of in vivo grown subgingival biofilm bacteria quantified by digital image analysis. PLoS ONE 7, e37583 (2012).
Settem, R. P., El-Hassan, A. T., Honma, K., Stafford, G. P. & Sharma, A. Fusobacterium nucleatum and Tannerella forsythia induce synergistic alveolar bone loss in a mouse periodontitis model. Infect. Immun. 80, 2436–2443 (2012).
Iwase, T. et al. Staphylococcus epidermidis Esp inhibits Staphylococcus aureus biofilm formation and nasal colonization. Nature 465, 346–349 (2010).
Fukuda, S. et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature 469, 543–547 (2011).
Guo, L. et al. Precision-guided antimicrobial peptide as a targeted modulator of human microbial ecology. Proc. Natl Acad. Sci. USA 112, 7569–7574 (2015).
Langermann, S. et al. Prevention of mucosal Escherichia coli infection by FimH-adhesin-based systemic vaccination. Science 276, 607–611 (1997).
Wessel, A. K., Hmelo, L., Parsek, M. R. & Whiteley, M. Going local: technologies for exploring bacterial microenvironments. Nat. Rev. Microbiol. 11, 337–348 (2013).
Leung, M. H., Wilkins, D. & Lee, P. K. Insights into the pan-microbiome: skin microbial communities of Chinese individuals differ from other racial groups. Sci. Rep. 5, 11845 (2015).
Gould, J. C. & Mc, K. E. The carriage of Staphylococcus pyogenes var. aureus in the human nose. J. Hyg. (Lond.) 52, 304–310 (1954).
Krismer, B. et al. Nutrient limitation governs Staphylococcus aureus metabolism and niche adaptation in the human nose. PLoS Pathog. 10, e1003862 (2014).
Jeraldo, P. et al. Quantification of the relative roles of niche and neutral processes in structuring gastrointestinal microbiomes. Proc. Natl Acad. Sci. USA 109, 9692–9698 (2012).
Rosindell, J., Hubbell, S. P. & Etienne, R. S. The unified neutral theory of biodiversity and biogeography at age ten. Trends Ecol. Evol. 26, 340–348 (2011).
Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 (2012).
Grice, E. A. et al. Topographical and temporal diversity of the human skin microbiome. Science 324, 1190–1192 (2009).
Dominguez-Bello, M. G. et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl Acad. Sci. USA 107, 11971–11975 (2010).
Koch, G. et al. Evolution of resistance to a last-resort antibiotic in Staphylococcus aureus via bacterial competition. Cell 158, 1060–1071 (2014).
Planet, P. J. et al. Emergence of the epidemic methicillin-resistant Staphylococcus aureus strain USA300 coincides with horizontal transfer of the arginine catabolic mobile element and speG-mediated adaptations for survival on skin. mBio 4, e00889-13 (2013).
de Wit, R. & Bouvier, T. 'Everything is everywhere, but, the environment selects'; what did Baas Becking and Beijerinck really say? Environ. Microbiol. 8, 755–758 (2006).
Finlay, B. J. Global dispersal of free-living microbial eukaryote species. Science 296, 1061–1063 (2002).
Whitaker, R. J., Grogan, D. W. & Taylor, J. W. Geographic barriers isolate endemic populations of hyperthermophilic archaea. Science 301, 976–978 (2003).
Martiny, J. B. et al. Microbial biogeography: putting microorganisms on the map. Nat. Rev. Microbiol. 4, 102–112 (2006).
Dowd, S. E. et al. Survey of bacterial diversity in chronic wounds using pyrosequencing, DGGE, and full ribosome shotgun sequencing. BMC Microbiol. 8, 43 (2008).
Griffin, A. S., West, S. A. & Buckling, A. Cooperation and competition in pathogenic bacteria. Nature 430, 1024–1027 (2004).
Diggle, S. P., Griffin, A. S., Campbell, G. S. & West, S. A. Cooperation and conflict in quorum-sensing bacterial populations. Nature 450, 411–414 (2007).
Kummerli, R., Gardner, A., West, S. A. & Griffin, A. S. Limited dispersal, budding dispersal, and cooperation: an experimental study. Evolution 63, 939–949 (2009).
Dimitriu, T. et al. Genetic information transfer promotes cooperation in bacteria. Proc. Natl Acad. Sci. USA 111, 11103–11108 (2014).
Schluter, J., Nadell, C. D., Bassler, B. L. & Foster, K. R. Adhesion as a weapon in microbial competition. ISME J. 9, 139–149 (2015).
Wang, M., Schaefer, A. L., Dandekar, A. A. & Greenberg, E. P. Quorum sensing and policing of Pseudomonas aeruginosa social cheaters. Proc. Natl Acad. Sci. USA 112, 2187–2191 (2015).
Rumbaugh, K. P. et al. Quorum sensing and the social evolution of bacterial virulence. Curr. Biol. 19, 341–345 (2009).
Arrhenius, O. Species and area. J. Ecol. 9, 95–99 (1921).
Gleason, H. On the relation between species and area. Ecology 3, 158–162 (1922).
Cornell, H. L., J. H. Species interactions, local and regional processes, and limits to the richness of ecological communities: a theoretical perspective. J. Anim. Ecol. 61, 1–12 (1992).
Lawton, J. Are there general laws in ecology? Oikos 84, 177–192 (1999).
Hillebrand, H. B., T. Regional and local impact on species diversity - from pattern to processes. Oecologia 132, 479–491 (2002).
Horner-Devine, M. C., Lage, M., Hughes, J. B. & Bohannan, B. J. A taxa-area relationship for bacteria. Nature 432, 750–753 (2004).
Bell, T. et al. Larger islands house more bacterial taxa. Science 308, 1884 (2005).
Ranjard, L. et al. Turnover of soil bacterial diversity driven by wide-scale environmental heterogeneity. Nat. Commun. 4, 1434 (2013).
Acknowledgements
The authors thank members of the Whiteley laboratory for critical discussion of this manuscript. This work was supported by a US National Institutes of Health (NIH) Grant 1R01DE020100 (to M.W.) and a Human Frontier Science Program (HFSP) grant HFSP RGP0011/2014 (to S.P.B. and M.W.). M.W. is a Burroughs Wellcome Investigator in the Pathogenesis of Infectious Disease program.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Glossary
- Biofilms
-
Surface-attached microbial communities that are encased in a matrix (for example, of polysaccharides, proteins, and/or DNA) and are often polymicrobial as well as highly resistant to antibiotic therapy and the host immune system.
- Polymicrobial
-
Diverse in species and/or strain content.
- Aggregate
-
A population of very few to several cells arranged in a cluster (otherwise known as a microcolony or in vivo biofilm) with many of the same properties that are observed in much larger, traditional biofilms studied in vitro.
- Synergy
-
A positive interaction term meaning 'greater than the sum of parts'. Here we use synergy to refer to microbial interactions that shape host health (disease synergy: two or more species in combination cause more severe infection than either could when acting alone) or microbial growth (growth synergy: each species grows better in combination than when alone). Note that growth synergy often, but not always, implies disease synergy.
- Metagenomics
-
Sequence-based analysis of the DNA recovered from microbial communities. These studies describe the genetic repertoire of a microbial community.
- Metatranscriptomics
-
Sequence-based analysis of the mRNA recovered from microbial communities. These studies provide information regarding gene expression in a microbial community.
- Microbiogeography
-
Although 'biogeography' refers to the distribution of species through space and time, 'microbiogeography', as defined in this Review, is the spatial organization of pathogen and commensal microbial populations at the scale of single infections.
- Pellicle
-
The permanent layer of proteins that coat oral surfaces and provide binding sites for early colonizers of dental plaque.
- Pioneer species
-
The first species to colonize previously disrupted ecosystems.
- Fimbriae
-
Hair-like bacterial appendages (also known as pili) that mediate surface attachment.
- Cystic fibrosis
-
A human genetic disorder in which a defect in a transmembrane ion channel causes the accumulation of mucus in the lungs, acting as a rich substrate for microbial growth.
- Quorum sensing
-
Density-dependent cell–cell communication where a constitutively produced signal, once it accumulates to a threshold concentration, can trigger microbial group behaviours.
- Type III secretion system
-
A needle-like bacterial apparatus that delivers effector proteins into host cells.
- Lactoferrin
-
A bactericidal host protein that sequesters iron.
- Mucin
-
A class of gel-forming proteins that give mucus its viscous property.
- Neutrophils
-
Host immune cells that unleash a mixture of redox-active molecules, in a process known as respiratory burst, to kill microorganisms.
- Aerotaxis
-
Chemotaxis in response to an oxygen gradient.
- Cheat
-
In social evolution, community members that exploit, but do not contribute to, the production of 'public goods'.
- Respiration
-
A metabolic growth process characterized by the reduction of an electron acceptor (such as oxygen or nitrate).
- Intestinal crypts
-
The narrow spaces that lie between villi (multicellular host structures that assist in nutrient absorption) in the small and large intestine that, in a non-diseased state, are very low in microbial presence.
- RegIIIγ
-
A host antimicrobial lectin (polysaccharide-binding protein) that targets Gram-positive bacteria.
- Mucin 2
-
(MUC2). A primary mucin (gel-forming glycoprotein) in the mucus layers of the small and large intestine.
- Co-aggregation
-
Intercellular binding between genetically distinct cells, often mediated by the recognition of a polysaccharide on the target cell by a cognate surface protein on the partner cell.
- Mucoid
-
A phenotypic variant that overproduces exopolysaccharide (for example, alginate produced by Pseudomonas aeruginosa).
- Autoinducer 2
-
A signalling molecule that is synthesized and sensed by many bacterial species.
- Pyocyanin
-
A broad-spectrum toxin produced by Pseudomonas aeruginosa that is upregulated in response to cell-wall fragments shed by Staphylococcus aureus, underlying synergistic virulence of these species in wound infections.
- Cross-feeding
-
The consumption of a waste product of one microorganism by another microorganism that can utilize the waste product as a nutrient source.
- Cross-protection
-
The shielding of one microorganism by another microorganism from an external stress.
- Probiotics
-
Microorganisms that promote host health.
Rights and permissions
About this article
Cite this article
Stacy, A., McNally, L., Darch, S. et al. The biogeography of polymicrobial infection. Nat Rev Microbiol 14, 93–105 (2016). https://doi.org/10.1038/nrmicro.2015.8
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrmicro.2015.8
This article is cited by
-
Formation of a biofilm matrix network shapes polymicrobial interactions
The ISME Journal (2023)
-
Chronic wounds
Nature Reviews Disease Primers (2022)
-
The biogeography of infection revisited
Nature Reviews Microbiology (2022)
-
Arc and pulsed spark discharge inactivation of pathogenic P. aeruginosa, S. aureus, M. canis, T. mentagrophytes, and C. albicans microorganisms
Environmental Science and Pollution Research (2022)
-
Droplet printing reveals the importance of micron-scale structure for bacterial ecology
Nature Communications (2021)