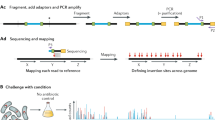
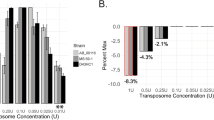
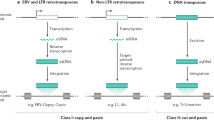
Abstract
Transposon insertion sequencing (TIS) is a powerful approach that can be extensively applied to the genome-wide definition of loci that are required for bacterial growth under diverse conditions. However, experimental design choices and stochastic biological processes can heavily influence the results of TIS experiments and affect downstream statistical analysis. In this Opinion article, we discuss TIS experimental parameters and how these factors relate to the benefits and limitations of the various statistical frameworks that can be applied to the computational analysis of TIS data.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Barquist, L., Boinett, C. J. & Cain, A. K. Approaches to querying bacterial genomes with transposon-insertion sequencing. RNA Biol. 10, 1161–1169 (2013).
van Opijnen, T. & Camilli, A. Transposon insertion sequencing: a new tool for systems-level analysis of microorganisms. Nat. Rev. Microbiol. 11, 435–442 (2013).
van Opijnen, T., Bodi, K. L. & Camilli, A. Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat. Methods 6, 767–772 (2009).
Goodman, A. L. et al. Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell Host Microbe 6, 279–289 (2009).
Gawronski, J. D., Wong, S. M., Giannoukos, G., Ward, D. V. & Akerley, B. J. Tracking insertion mutants within libraries by deep sequencing and a genome-wide screen for Haemophilus genes required in the lung. Proc. Natl Acad. Sci. USA 106, 16422–16427 (2009).
Langridge, G. C. et al. Simultaneous assay of every Salmonella Typhi gene using one million transposon mutants. Genome Res. 19, 2308–2316 (2009).
Chiang, S. L. & Rubin, E. J. Construction of a mariner-based transposon for epitope-tagging and genomic targeting. Gene 296, 179–185 (2002).
Rubin, E. J. et al. In vivo transposition of mariner-based elements in enteric bacteria and mycobacteria. Proc. Natl Acad. Sci. USA 96, 1645–1650 (1999).
Goryshin, I. Y., Miller, J. A., Kil, Y. V., Lanzov, V. A. & Reznikoff, W. S. Tn5/IS50 target recognition. Proc. Natl Acad. Sci. USA 95, 10716–10721 (1998).
Zhang, Y. J. et al. Global assessment of genomic regions required for growth in Mycobacterium tuberculosis. PLoS Pathog. 8, e1002946 (2012).
Chao, M. C. et al. High-resolution definition of the Vibrio cholerae essential gene set with hidden Markov model-based analyses of transposon-insertion sequencing data. Nucleic Acids Res. 41, 9033–9048 (2013).
Lodge, J. K., Weston-Hafer, K. & Berg, D. E. Transposon Tn5 target specificity: preference for insertion at G/C pairs. Genetics 120, 645–650 (1988).
Green, B., Bouchier, C., Fairhead, C., Craig, N. L. & Cormack, B. P. Insertion site preference of Mu, Tn5, and Tn7 transposons. Mob. DNA 3, 3 (2012).
Christen, B. et al. The essential genome of a bacterium. Mol. Syst. Biol. 7, 528 (2011).
Barquist, L. et al. A comparison of dense transposon insertion libraries in the Salmonella serovars Typhi and Typhimurium. Nucleic Acids Res. 41, 4549–4564 (2013).
DeJesus, M. A. et al. Bayesian analysis of gene essentiality based on sequencing of transposon insertion libraries. Bioinformatics 29, 695–703 (2013).
Lamichhane, G. et al. A postgenomic method for predicting essential genes at subsaturation levels of mutagenesis: application to Mycobacterium tuberculosis. Proc. Natl Acad. Sci. USA 100, 7213–7218 (2003).
Griffin, J. E. et al. High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PLoS Pathog. 7, e1002251 (2011).
Zemansky, J. et al. Development of a mariner-based transposon and identification of Listeria monocytogenes determinants, including the peptidyl-prolyl isomerase PrsA2, that contribute to its hemolytic phenotype. J. Bacteriol. 191, 3950–3964 (2009).
Johnson, C. M. & Grossman, A. D. Identification of host genes that affect acquisition of an integrative and conjugative element in Bacillus subtilis. Mol. Microbiol. 93, 1284–1301 (2014).
Shevchenko, Y. et al. Systematic sequencing of cDNA clones using the transposon Tn5. Nucleic Acids Res. 30, 2469–2477 (2002).
Troy, E. B. et al. Understanding barriers to Borrelia burgdorferi dissemination during infection using massively parallel sequencing. Infect. Immun. 81, 2347–2357 (2013).
Fu, Y., Waldor, M. K. & Mekalanos, J. J. Tn-Seq analysis of Vibrio cholerae intestinal colonization reveals a role for T6SS-mediated antibacterial activity in the host. Cell Host Microbe 14, 652–663 (2013).
Pritchard, J. R. et al. ARTIST: high-resolution genome-wide assessment of fitness using transposon-insertion sequencing. PLoS Genet. 10, e1004782 (2014).
van Opijnen, T. & Camilli, A. A fine scale phenotype-genotype virulence map of a bacterial pathogen. Genome Res. 22, 2541–2551 (2012).
Carter, R. et al. Genomic analyses of pneumococci from children with sickle cell disease expose host-specific bacterial adaptations and deficits in current interventions. Cell Host Microbe 15, 587–599 (2014).
Abel, S. et al. Sequence tag-based analysis of microbial population dynamics. Nat. Methods 12, 223–226 (2015).
Kaper, J. B., Morris, J. G. Jr & Levine, M. M. Cholera. Clin. Microbiol. Rev. 8, 48–86 (1995).
Ritchie, J. M., Rui, H., Bronson, R. T. & Waldor, M. K. Back to the future: studying cholera pathogenesis using infant rabbits. mBio 1, e00047-10 (2010).
Kamp, H. D., Patimalla-Dipali, B., Lazinski, D. W., Wallace-Gadsden, F. & Camilli, A. Gene fitness landscapes of Vibrio cholerae at important stages of its life cycle. PLoS Pathog. 9, e1003800 (2013).
Zomer, A., Burghout, P., Bootsma, H. J., Hermans, P. W. & van Hijum, S. A. ESSENTIALS: software for rapid analysis of high throughput transposon insertion sequencing data. PLoS ONE 7, e43012 (2012).
Lazinski, D. W. & Camilli, A. Homopolymer tail-mediated ligation PCR: a streamlined and highly efficient method for DNA cloning and library construction. Biotechniques 54, 25–34 (2013).
Gallagher, L. A., Shendure, J. & Manoil, C. Genome-scale identification of resistance functions in Pseudomonas aeruginosa using Tn-seq. mBio 2, e00315-10 (2011).
Yamaichi, Y. et al. High-resolution genetic analysis of the requirements for horizontal transmission of the ESBL plasmid from Escherichia coli O104:H4. Nucleic Acids Res. 43, 348–360 (2014).
Fu, G. K. et al. Molecular indexing enables quantitative targeted RNA sequencing and reveals poor efficiencies in standard library preparations. Proc. Natl Acad. Sci. USA 111, 1891–1896 (2014).
Shiroguchi, K., Jia, T. Z., Sims, P. A. & Xie, X. S. Digital RNA sequencing minimizes sequence-dependent bias and amplification noise with optimized single-molecule barcodes. Proc. Natl Acad. Sci. USA 109, 1347–1352 (2012).
Li, H. et al. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Baugh, L. et al. Combining functional and structural genomics to sample the essential Burkholderia structome. PLoS ONE 8, e53851 (2013).
Valentino, M. D. et al. Genes contributing to Staphylococcus aureus fitness in abscess- and infection-related ecologies. mBio 5, e01729-14 (2014).
Klein, B. A. et al. Identification of essential genes of the periodontal pathogen Porphyromonas gingivalis. BMC Genomics 13, 578 (2012).
Deng, J., Su, S., Lin, X., Hassett, D. J. & Lu, L. J. A statistical framework for improving genomic annotations of prokaryotic essential genes. PLoS ONE 8, e58178 (2013).
Remmele, C. W. et al. Transcriptional landscape and essential genes of Neisseria gonorrhoeae. Nucleic Acids Res. 42, 10579–10595 (2014).
DeJesus, M. A. & Ioerger, T. R. A. Hidden Markov Model for identifying essential and growth-defect regions in bacterial genomes from transposon insertion sequencing data. BMC Bioinformatics 14, 303 (2013).
Brutinel, E. D. & Gralnick, J. A. Anomalies of the anaerobic tricarboxylic acid cycle in Shewanella oneidensis revealed by Tn-seq. Mol. Microbiol. 86, 273–283 (2012).
Zhang, Y. J. et al. Tryptophan biosynthesis protects mycobacteria from CD4 T-cell-mediated killing. Cell 155, 1296–1308 (2013).
Mann, B. et al. Control of virulence by small RNAs in Streptococcus pneumoniae. PLoS Pathog. 8, e1002788 (2012).
McDonough, E., Lazinski, D. W. & Camilli, A. Identification of in vivo regulators of the Vibrio cholerae xds gene using a high-throughput genetic selection. Mol. Microbiol. 92, 302–315 (2014).
Moll, A. et al. Cell separation in Vibrio cholerae is mediated by a single amidase whose action is modulated by two nonredundant activators. J. Bacteriol. 196, 3937–3948 (2014).
Dorr, T. et al. A novel peptidoglycan binding protein crucial for PBP1A-mediated cell wall biogenesis in Vibrio cholerae. PLoS Genet. 10, e1004433 (2014).
Skurnik, D. et al. A comprehensive analysis of in vitro and in vivo genetic fitness of Pseudomonas aeruginosa using high-throughput sequencing of transposon libraries. PLoS Pathog. 9, e1003582 (2013).
Turner, K. H., Everett, J., Trivedi, U., Rumbaugh, K. P. & Whiteley, M. Requirements for Pseudomonas aeruginosa acute burn and chronic surgical wound infection. PLoS Genet. 10, e1004518 (2014).
Wang, Z., Gerstein, M. & Snyder, M. RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 10, 57–63 (2009).
Trapnell, C. et al. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 31, 46–53 (2013).
Anders, S. & Huber, W. Differential expression analysis for sequence count data. Genome Biol. 11, R106 (2010).
Khatiwara, A. et al. Genome scanning for conditionally essential genes in Salmonella enterica serotype Typhimurium. Appl. Environ. Microbiol. 78, 3098–3107 (2012).
Santa Maria, J. P. Jr et al. Compound-gene interaction mapping reveals distinct roles for Staphylococcus aureus teichoic acids. Proc. Natl Acad. Sci. USA 111, 12510–12515 (2014).
Hsiao, A. et al. Members of the human gut microbiota involved in recovery from Vibrio cholerae infection. Nature 515, 423–426 (2014).
Moxon, E. R. & Murphy, P. A. Haemophilus influenzae bacteremia and meningitis resulting from survival of a single organism. Proc. Natl Acad. Sci. USA 75, 1534–1536 (1978).
Barnes, P. D., Bergman, M. A., Mecsas, J. & Isberg, R. R. Yersinia pseudotuberculosis disseminates directly from a replicating bacterial pool in the intestine. J. Exp. Med. 203, 1591–1601 (2006).
Kaiser, P., Slack, E., Grant, A. J., Hardt, W. D. & Regoes, R. R. Lymph node colonization dynamics after oral Salmonella typhimurium infection in mice. PLoS Pathog. 9, e1003532 (2013).
Grant, A. J. et al. Modelling within-host spatiotemporal dynamics of invasive bacterial disease. PLoS Biol. 6, e74 (2008).
Acknowledgements
This work was supported by the Howard Hughes Medical Institute, the US National Institutes of Health (AI R37-042347 to M.K.W.; 5F32 GM108355-02 to M.C.C.), and the Swiss Foundation for Grants in Biology and Medicine (PASMP3_142724/1 to S.A.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
PowerPoint slides
Rights and permissions
About this article
Cite this article
Chao, M., Abel, S., Davis, B. et al. The design and analysis of transposon insertion sequencing experiments. Nat Rev Microbiol 14, 119–128 (2016). https://doi.org/10.1038/nrmicro.2015.7
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrmicro.2015.7
This article is cited by
-
Functional annotation and importance of marine bacterial transporters of plankton exometabolites
ISME Communications (2023)
-
Mutagenesis reveals how Akkermansia muciniphila degrades mucin and colonizes the gut
Nature Microbiology (2023)
-
Identification of determinants for entering into a viable but nonculturable state in Vibrio alginolyticus by Tn-seq
Applied Microbiology and Biotechnology (2023)
-
Long-read sequencing for identification of insertion sites in large transposon mutant libraries
Scientific Reports (2022)
-
Identification of putative essential protein domains from high-density transposon insertion sequencing
Scientific Reports (2022)