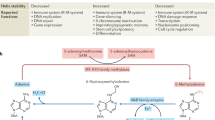
Abstract
DNA N6-adenine methylation (N6-methyladenine; 6mA) in prokaryotes functions primarily in the host defence system. The prevalence and significance of this modification in eukaryotes had been unclear until recently. Here, we discuss recent publications documenting the presence of 6mA in Chlamydomonas reinhardtii, Drosophila melanogaster and Caenorhabditis elegans; consider possible roles for this DNA modification in regulating transcription, the activity of transposable elements and transgenerational epigenetic inheritance; and propose 6mA as a new epigenetic mark in eukaryotes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Vanyushin, B. F., Tkacheva, S. G. & Belozersky, A. N. Rare bases in animal DNA. Nature 225, 948–949 (1970).
Dunn, D. B. & Smith, J. D. Occurrence of a new base in the deoxyribonucleic acid of a strain of Bacterium coli. Nature 175, 336–337 (1955).
Vanyushin, B. F., Belozersky, A. N., Kokurina, N. A. & Kadirova, D. X. 5-methylcytosine and 6-methylamino-purine in bacterial DNA. Nature 218, 1066–1067 (1968).
Unger, G. & Venner, H. Remarks on minor bases in spermatic desoxyribonucleic acid. Hoppe Seylers Z. Physiol. Chem. 344, 280–283 (1966).
Hattman, S., Kenny, C., Berger, L. & Pratt, K. Comparative study of DNA methylation in three unicellular eucaryotes. J. Bacteriol. 135, 1156–1157 (1978).
Feng, S. H. et al. Conservation and divergence of methylation patterning in plants and animals. Proc. Natl Acad. Sci. USA 107, 8689–8694 (2010).
Smith, Z. D. & Meissner, A. DNA methylation: roles in mammalian development. Nat. Rev. Genet. 14, 204–220 (2013).
Jones, P. A. & Takai, D. The role of DNA methylation in mammalian epigenetics. Science 293, 1068–1070 (2001).
Jones, P. A. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 13, 484–492 (2012).
Naito, T., Kusano, K. & Kobayashi, I. Selfish behavior of restriction–modification systems. Science 267, 897–899 (1995).
Wion, D. & Casadesus, J. N6-methyl-adenine: an epigenetic signal for DNA–protein interactions. Nat. Rev. Microbiol. 4, 183–192 (2006).
Vasu, K. & Nagaraja, V. Diverse functions of restriction–modification systems in addition to cellular defense. Microbiol. Mol. Biol. Rev. 77, 53–72 (2013).
Urieli-Shoval, S., Gruenbaum, Y. & Razin, A. Sequence and substrate specificity of isolated DNA methylases from Escherichia coli C. J. Bacteriol. 153, 274–280 (1983).
Wright, R., Stephens, C. & Shapiro, L. The CcrM DNA methyltransferase is widespread in the alpha subdivision of proteobacteria, and its essential functions are conserved in Rhizobium meliloti and Caulobacter crescentus. J. Bacteriol. 179, 5869–5877 (1997).
Greer, E. L. et al. DNA methylation on N6-adenine in C. elegans. Cell 161, 868–878 (2015).
Zhang, G. et al. N6-methyladenine DNA modification in Drosophila. Cell 161, 893–906 (2015).
Fu, Y. et al. N6-methyldeoxyadenosine marks active transcription start sites in Chlamydomonas. Cell 161, 879–892 (2015).
Gorovsky, M. A., Hattman, S. & Pleger, G. L. [6N]methyl adenine in the nuclear DNA of a eucaryote, Tetrahymena pyriformis. J. Cell Biol. 56, 697–701 (1973).
Cummings, D. J., Tait, A. & Goddard, J. M. Methylated bases in DNA from Paramecium aurelia. Biochim. Biophys. Acta 374, 1–11 (1974).
Hattman, S. DNA-[adenine] methylation in lower eukaryotes. Biochemistry 70, 550–558 (2005).
Ratel, D., Ravanat, J. L., Berger, F. & Wion, D. N6-methyladenine: the other methylated base of DNA. Bioessays 28, 309–315 (2006).
Suzuki, M. M. & Bird, A. DNA methylation landscapes: provocative insights from epigenomics. Nat. Rev. Genet. 9, 465–476 (2008).
Adams, R. L., McKay, E. L., Craig, L. M. & Burdon, R. H. Methylation of mosquito DNA. Biochim. Biophys. Acta 563, 72–81 (1979).
Ashapkin, V. V., Kutueva, L. I. & Vanyushin, B. F. The gene for domains rearranged methyltransferase (DRM2) in Arabidopsis thaliana plants is methylated at both cytosine and adenine residues. FEBS Lett. 532, 367–372 (2002).
Vanyushin, B. F., Alexandrushkina, N. I. & Kirnos, M. D. N6-methyladenine in mitochondrial-DNA of higher-plants. Febs Lett. 233, 397–399 (1988).
Kay, P. H. et al. Evidence for adenine methylation within the mouse myogenic gene Myo-D1. Gene 151, 89–95 (1994).
Huang, W. et al. Determination of DNA adenine methylation in genomes of mammals and plants by liquid chromatography/mass spectrometry. RSC Adv. 5, 64046–64054 (2015).
Jia, G. et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 7, 885–887 (2011).
Dominissini, D. et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206 (2012).
Luo, G. Z. et al. Unique features of the m6A methylome in Arabidopsis thaliana. Nat. Commun. 5, 5630 (2014).
Frelon, S. et al. High-performance liquid chromatography—tandem mass spectrometry measurement of radiation-induced base damage to isolated and cellular DNA. Chem. Res. Toxicol. 13, 1002–1010 (2000).
Roberts, R. J. & Macelis, D. REBASE — restriction enzymes and methylases. Nucleic Acids Res. 29, 268–269 (2001).
Lacks, S. & Greenberg, B. Complementary specificity of restriction endonucleases of Diplococcus pneumoniae with respect to DNA methylation. J. Mol. Biol. 114, 153–168 (1977).
Laird, P. W. Principles and challenges of genomewide DNA methylation analysis. Nat. Rev. Genet. 11, 191–203 (2010).
Flusberg, B. A. et al. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat. Methods 7, 461–465 (2010).
Schadt, E. E. et al. Modeling kinetic rate variation in third generation DNA sequencing data to detect putative modifications to DNA bases. Genome Res. 23, 129–141 (2013).
Fang, G. et al. Genome-wide mapping of methylated adenine residues in pathogenic Escherichia coli using single-molecule real-time sequencing. Nat. Biotechnol. 30, 1232–1239 (2012).
Jones, P. L. et al. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat. Genet. 19, 187–191 (1998).
Touzain, F., Petit, M. A., Schbath, S. & El Karoui, M. DNA motifs that sculpt the bacterial chromosome. Nat. Rev. Microbiol. 9, 15–26 (2011).
Karrer, K. M. & VanNuland, T. A. Methylation of adenine in the nuclear DNA of Tetrahymena is internucleosomal and independent of histone H1. Nucleic Acids Res. 30, 1364–1370 (2002).
Bromberg, S., Pratt, K. & Hattman, S. Sequence specificity of DNA adenine methylase in the protozoan Tetrahymena thermophila. J. Bacteriol. 150, 993–996 (1982).
Fu, Y., Dominissini, D., Rechavi, G. & He, C. Gene expression regulation mediated through reversible m6A RNA methylation. Nat. Rev. Genet. 15, 293–306 (2014).
Jia, G. F. et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 7, 885–887 (2011).
Liu, J. et al. A METTL3–METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 10, 93–95 (2014).
Zheng, G. Q. et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 49, 18–29 (2013).
Iyer, L. M., Abhiman, S. & Aravind, L. Natural history of eukaryotic DNA methylation systems. Prog. Mol. Biol. Transl. Sci. 101, 25–104 (2011).
Song, J., Rechkoblit, O., Bestor, T. H. & Patel, D. J. Structure of DNMT1–DNA complex reveals a role for autoinhibition in maintenance DNA methylation. Science 331, 1036–1040 (2011).
Bokar, J. A., Shambaugh, M. E., Polayes, D., Matera, A. G. & Rottman, F. M. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA 3, 1233–1247 (1997).
Trewick, S. C., Henshaw, T. F., Hausinger, R. P., Lindahl, T. & Sedgwick, B. Oxidative demethylation by Escherichia coli AlkB directly reverts DNA base damage. Nature 419, 174–178 (2002).
Tahiliani, M. et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324, 930–935 (2009).
Yang, C. G. et al. Crystal structures of DNA/RNA repair enzymes AlkB and ABH2 bound to dsDNA. Nature 452, 961–965 (2008).
Hu, L. et al. Crystal structure of TET2–DNA complex: insight into TET-mediated 5mC oxidation. Cell 155, 1545–1555 (2013).
Bang, J., Bae, S. H., Park, C. J., Lee, J. H. & Choi, B. S. Structural and dynamics study of DNA dodecamer duplexes that contain un-, hemi-, or fully methylated GATC sites. J. Am. Chem. Soc. 130, 17688–17696 (2008).
Tronche, F., Rollier, A., Bach, I., Weiss, M. C. & Yaniv, M. The rat albumin promoter: cooperation with upstream elements is required when binding of APF/HNF1 to the proximal element is partially impaired by mutation or bacterial methylation. Mol. Cell. Biol. 9, 4759–4766 (1989).
Sugimoto, K., Takeda, S. & Hirochika, H. Transcriptional activation mediated by binding of a plant GATA-type zinc finger protein AGP1 to the AG-motif (AGATCCAA) of the wound-inducible Myb gene NtMyb2. Plant J. 36, 550–564 (2003).
Lichtsteiner, S. & Schibler, U. A glycosylated liver-specific transcription factor stimulates transcription of the albumin gene. Cell 57, 1179–1187 (1989).
Ratel, D. et al. The bacterial nucleoside N6-methyldeoxyadenosine induces the differentiation of mammalian tumor cells. Biochem. Biophys. Res. Commun. 285, 800–805 (2001).
van Blokland, R., Ross, S., Corrado, G., Scollan, C. & Meyer, P. Developmental abnormalities associated with deoxyadenosine methylation in transgenic tobacco. Plant J. 15, 543–551 (1998).
Roberts, D., Hoopes, B. C., Mcclure, W. R. & Kleckner, N. Is10 transposition is regulated by DNA adenine methylation. Cell 43, 117–130 (1985).
Hernday, A., Krabbe, M., Braaten, B. & Low, D. Self-perpetuating epigenetic pili switches in bacteria. Proc. Natl Acad. Sci. USA 99, 16470–16476 (2002).
Rogers, J. C. & Rogers, S. W. Comparison of the effects of N6-methyldeoxyadenosine and N5-methyldeoxycytosine on transcription from nuclear gene promoters in barley. Plant J. 7, 221–233 (1995).
Pratt, K. & Hattman, S. Deoxyribonucleic-acid methylation and chromatin organization in Tetrahymena thermophila. Mol. Cell. Biol. 1, 600–608 (1981).
Huff, J. T. & Zilberman, D. Dnmt1-independent CG methylation contributes to nucleosome positioning in diverse eukaryotes. Cell 156, 1286–1297 (2014).
Katz, D. J., Edwards, T. M., Reinke, V. & Kelly, W. G. A C. elegans LSD1 demethylase contributes to germline immortality by reprogramming epigenetic memory. Cell 137, 308–320 (2009).
Greer, E. L. et al. A histone methylation network regulates transgenerational epigenetic memory in C. elegans. Cell Rep. 7, 113–126 (2014).
Poole, A., Penny, D. & Sjoberg, B. M. Confounded cytosine! Tinkering and the evolution of DNA. Nat. Rev. Mol. Cell Biol. 2, 147–151 (2001).
Kierzek, E. & Kierzek, R. The thermodynamic stability of RNA duplexes and hairpins containing N6-alkyladenosines and 2-methylthio-N6-alkyladenosines. Nucleic Acids Res. 31, 4472–4480 (2003).
Roost, C. et al. Structure and thermodynamics of N6-methyladenosine in RNA: a spring-loaded base modification. J. Am. Chem. Soc. 137, 2107–2115 (2015).
Liu, N. et al. N6-methyladenosine-dependent RNA structural switches regulate RNA–protein interactions. Nature 518, 560–564 (2015).
Fu, Y. et al. FTO-mediated formation of N6-hydroxymethyladenosine and N6-formyladenosine in mammalian RNA. Nat. Commun. 4, 1798 (2013).
Capuano, F., Mulleder, M., Kok, R., Blom, H. J. & Ralser, M. Cytosine DNA methylation is found in Drosophila melanogaster but absent in Saccharomyces cerevisiae, Schizosaccharomyces pombe, and other yeast species. Anal. Chem. 86, 3697–3702 (2014).
Acknowledgements
We apologize to colleagues whose work was not cited owing to space limitations. C.H. is supported by the US National Institutes of Health (NIH) grant HG006827 and is an investigator of Howard Hughes Medical Institute. M.A.B. is a Special Fellow of the Leukemia & Lymphoma Society. Work in the Greer lab is supported by a National Institute on Aging of the NIH grant (AG043550). Work in the Shi laboratory is supported by grants from the NIH (CA118487 and MH096066), the Ellison Medical Foundation and the Samuel Waxman Cancer Research Foundation. Y.S. is an American Cancer Society Research Professor.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
Y.S. is a co-founder of Constellation Pharmaceuticals, Inc. and a member of its scientific advisory board. The other authors declare no competing interests.
Rights and permissions
About this article
Cite this article
Luo, GZ., Blanco, M., Greer, E. et al. DNA N6-methyladenine: a new epigenetic mark in eukaryotes?. Nat Rev Mol Cell Biol 16, 705–710 (2015). https://doi.org/10.1038/nrm4076
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrm4076
This article is cited by
-
6mA-METL-9 axis regulates innate immunity in C. elegans
Cell Research (2023)
-
Long range inter-chromosomal interaction of Oct4 distal enhancer loci regulates ESCs pluripotency
Cell Death Discovery (2023)
-
Hyb4mC: a hybrid DNA2vec-based model for DNA N4-methylcytosine sites prediction
BMC Bioinformatics (2022)
-
Bacterial N4-methylcytosine as an epigenetic mark in eukaryotic DNA
Nature Communications (2022)