Abstract
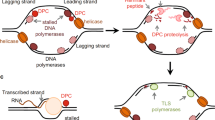
DNA–protein crosslinks (DPCs) are highly toxic DNA adducts, but whether dedicated DPC-repair mechanisms exist was until recently unknown. This has changed with discoveries made in yeast and Xenopus laevis that revealed a protease-based DNA-repair pathway specific for DPCs. Importantly, mutations in the gene encoding the putative human homologue of a yeast DPC protease cause a human premature ageing and cancer predisposition syndrome. Thus, DPC repair is a previously overlooked genome-maintenance mechanism that may be essential for tumour suppression.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Hoeijmakers, J. H. DNA damage, aging, and cancer. N. Engl. J. Med. 361, 1475–1485 (2009).
Jackson, S. P. & Bartek, J. The DNA-damage response in human biology and disease. Nature 461, 1071–1078 (2009).
Barker, S., Weinfeld, M. & Murray, D. DNA–protein crosslinks: their induction, repair, and biological consequences. Mutat. Res. 589, 111–135 (2005).
Stingele, J., Schwarz, M. S., Bloemeke, N., Wolf, P. G. & Jentsch, S. A. DNA-dependent protease involved in DNA-protein crosslink repair. Cell 158, 327–338 (2014).
Duxin, J. P., Dewar, J. M., Yardimci, H. & Walter, J. C. Repair of a DNA-protein crosslink by replication-coupled proteolysis. Cell 159, 346–357 (2014).
Kiianitsa, K. & Maizels, N. A rapid and sensitive assay for DNA–protein covalent complexes in living cells. Nucleic Acids Res. 41, e104 (2013).
Chen, S. H., Chan, N. L. & Hsieh, T. S. New mechanistic and functional insights into DNA topoisomerases. Annu. Rev. Biochem. 82, 139–170 (2013).
Pommier, Y. Topoisomerase I inhibitors: camptothecins and beyond. Nat. Rev. Cancer 6, 789–802 (2006).
Pourquier, P. et al. Effects of uracil incorporation, DNA mismatches, and abasic sites on cleavage and religation activities of mammalian topoisomerase I. J. Biol. Chem. 272, 7792–7796 (1997).
Pommier, Y. et al. Tyrosyl-DNA-phosphodiesterases (TDP1 and TDP2). DNA Repair 19, 114–129 (2014).
Shi, Y. et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119, 941–953 (2004).
Tsukada, Y. et al. Histone demethylation by a family of JmjC domain-containing proteins. Nature 439, 811–816 (2006).
Shen, L., Song, C. X., He, C. & Zhang, Y. Mechanism and function of oxidative reversal of DNA and RNA methylation. Annu. Rev. Biochem. 83, 585–614 (2014).
Trewick, S. C., Henshaw, T. F., Hausinger, R. P., Lindahl, T. & Sedgwick, B. Oxidative demethylation by Escherichia coli AlkB directly reverts DNA base damage. Nature 419, 174–178 (2002).
Solomon, M. J., Larsen, P. L. & Varshavsky, A. Mapping protein-DNA interactions in vivo with formaldehyde: evidence that histone H4 is retained on a highly transcribed gene. Cell 53, 937–947 (1988).
Lu, K. et al. Structural characterization of formaldehyde-induced cross-links between amino acids and deoxynucleosides and their oligomers. J. Am. Chem. Soc. 132, 3388–3399 (2010).
Ward, J. F. The complexity of DNA damage: relevance to biological consequences. Int. J. Radiat. Biol. 66, 427–432 (1994).
Zhang, H., Koch, C. J., Wallen, C. A. & Wheeler, K. T. Radiation-induced DNA damage in tumors and normal tissues. III. Oxygen dependence of the formation of strand breaks and DNA-protein crosslinks. Radiat. Res. 142, 163–168 (1995).
Fornace, A. J. Jr & Little, J. B. DNA crosslinking induced by X-rays and chemical agents. Biochim. Biophys. Acta 477, 343–355 (1977).
Shoulkamy, M. I. et al. Detection of DNA–protein crosslinks (DPCs) by novel direct fluorescence labeling methods: distinct stabilities of aldehyde and radiation-induced DPCs. Nucleic Acids Res. 40, e143 (2012).
Nitiss, J. L. Targeting DNA topoisomerase II in cancer chemotherapy. Nat. Rev. Cancer 9, 338–350 (2009).
Nakano, T. et al. T7 RNA polymerases backed up by covalently trapped proteins catalyze highly error prone transcription. J. Biol. Chem. 287, 6562–6572 (2012).
Nakano, T. et al. Translocation and stability of replicative DNA helicases upon encountering DNA–protein cross-links. J. Biol. Chem. 288, 4649–4658 (2013).
Fu, Y. V. et al. Selective bypass of a lagging strand roadblock by the eukaryotic replicative DNA helicase. Cell 146, 931–941 (2011).
Kuo, H. K., Griffith, J. D. & Kreuzer, K. N. 5-azacytidine induced methyltransferase-DNA adducts block DNA replication in vivo. Cancer Res. 67, 8248–8254 (2007).
Yeo, J. E. et al. Synthesis of site-specific DNA-protein conjugates and their effects on DNA replication. ACS Chem. Biol. 9, 1860–1868 (2014).
Novakova, O., Kasparkova, J., Malina, J., Natile, G. & Brabec, V. DNA–protein cross-linking by trans-[PtCl2(E-iminoether)2]. A concept for activation of the trans geometry in platinum antitumor complexes. Nucleic Acids Res. 31, 6450–6460 (2003).
Chvalova, K., Brabec, V. & Kasparkova, J. Mechanism of the formation of DNA–protein cross-links by antitumor cisplatin. Nucleic Acids Res. 35, 1812–1821 (2007).
Titenko-Holland, N. et al. Quantification of epithelial cell micronuclei by fluorescence in situ hybridization (FISH) in mortuary science students exposed to formaldehyde. Mutat. Res. 371, 237–248 (1996).
Basler, A., v d Hude, W. & Scheutwinkel-Reich, M. Formaldehyde-induced sister chromatid exchanges in vitro and the influence of the exogenous metabolizing systems S9 mix and primary rat hepatocytes. Arch. Toxicol. 58, 10–13 (1985).
Grogan, D. & Jinks-Robertson, S. Formaldehyde-induced mutagenesis in Saccharomyces cerevisiae: molecular properties and the roles of repair and bypass systems. Mutat. Res. 731, 92–98 (2012).
Swenberg, J. A. et al. Endogenous versus exogenous DNA adducts: their role in carcinogenesis, epidemiology, and risk assessment. Toxicol. Sci. 120, S130–S145 (2011).
Hartsuiker, E., Neale, M. J. & Carr, A. M. Distinct requirements for the Rad32Mre11 nuclease and Ctp1CtIP in the removal of covalently bound topoisomerase I and II from DNA. Mol. Cell 33, 117–123 (2009).
Hartsuiker, E. et al. Ctp1CtIP and Rad32Mre11 nuclease activity are required for Rec12Spo11 removal, but Rec12Spo11 removal is dispensable for other MRN-dependent meiotic functions. Mol. Cell. Biol. 29, 1671–1681 (2009).
Cannavo, E. & Cejka, P. Sae2 promotes dsDNA endonuclease activity within Mre11-Rad50-Xrs2 to resect DNA breaks. Nature 514, 122–125 (2014).
de Graaf, B., Clore, A. & McCullough, A. K. Cellular pathways for DNA repair and damage tolerance of formaldehyde-induced DNA-protein crosslinks. DNA Repair 8, 1207–1214 (2009).
Nakano, T. et al. Homologous recombination but not nucleotide excision repair plays a pivotal role in tolerance of DNA-protein cross-links in mammalian cells. J. Biol. Chem. 284, 27065–27076 (2009).
Nakano, T. et al. Nucleotide excision repair and homologous recombination systems commit differentially to the repair of DNA-protein crosslinks. Mol. Cell 28, 147–158 (2007).
Fornace, A. J. Jr. Detection of DNA single-strand breaks produced during the repair of damage by DNA-protein cross-linking agents. Cancer Res. 42, 145–149 (1982).
Baker, D. J. et al. Nucleotide excision repair eliminates unique DNA-protein cross-links from mammalian cells. J. Biol. Chem. 282, 22592–22604 (2007).
Quievryn, G. & Zhitkovich, A. Loss of DNA–protein crosslinks from formaldehyde-exposed cells occurs through spontaneous hydrolysis and an active repair process linked to proteosome function. Carcinogenesis 21, 1573–1580 (2000).
Ridpath, J. R. et al. Cells deficient in the FANC/BRCA pathway are hypersensitive to plasma levels of formaldehyde. Cancer Res. 67, 11117–11122 (2007).
Orta, M. L. et al. 5-Aza-2′-deoxycytidine causes replication lesions that require Fanconi anemia-dependent homologous recombination for repair. Nucleic Acids Res. 41, 5827–5836 (2013).
Rosado, I. V., Langevin, F., Crossan, G. P., Takata, M. & Patel, K. J. Formaldehyde catabolism is essential in cells deficient for the Fanconi anemia DNA-repair pathway. Nat. Struct. Mol. Biol. 18, 1432–1434 (2011).
Langevin, F., Crossan, G. P., Rosado, I. V., Arends, M. J. & Patel, K. J. Fancd2 counteracts the toxic effects of naturally produced aldehydes in mice. Nature 475, 53–58 (2011).
Garaycoechea, J. I. et al. Genotoxic consequences of endogenous aldehydes on mouse haematopoietic stem cell function. Nature 489, 571–575 (2012).
Stingele, J., Habermann, B. & Jentsch, S. DNA–protein crosslink repair: proteases as DNA repair enzymes. Trends Biochem. Sci. 40, 67–71 (2015).
Centore, R. C., Yazinski, S. A., Tse, A. & Zou, L. Spartan/C1orf124, a reader of PCNA ubiquitylation and a regulator of UV-induced DNA damage response. Mol. Cell 46, 625–635 (2012).
Fang, Q. DNA–protein crosslinks processed by nucleotide excision repair and homologous recombination with base and strand preference in E. coli model system. Mutat. Res. 741–742, 1–10 (2013).
Davis, E. J. et al. DVC1 (C1orf124) recruits the p97 protein segregase to sites of DNA damage. Nat. Struct. Mol. Biol. 19, 1093–1100 (2012).
Juhasz, S. et al. Characterization of human Spartan/C1orf124, an ubiquitin-PCNA interacting regulator of DNA damage tolerance. Nucleic Acids Res. 40, 10795–10808 (2012).
Machida, Y., Kim, M. S. & Machida, Y. J. Spartan/C1orf124 is important to prevent UV-induced mutagenesis. Cell Cycle 11, 3395–3402 (2012).
Ghosal, G., Leung, J. W., Nair, B. C., Fong, K. W. & Chen, J. Proliferating cell nuclear antigen (PCNA)-binding protein C1orf124 is a regulator of translesion synthesis. J. Biol. Chem. 287, 34225–34233 (2012).
Delabaere, L. et al. The spartan ortholog maternal haploid is required for paternal chromosome integrity in the Drosophila zygote. Curr. Biol. 24, 2281–2287 (2014).
Lessel, D. et al. Mutations in SPRTN cause early onset hepatocellular carcinoma, genomic instability and progeroid features. Nat. Genet. 46, 1239–1244 (2014).
Maskey, R. S. et al. Spartan deficiency causes genomic instability and progeroid phenotypes. Nat. Commun. 5, 5744 (2014).
Acknowledgements
S.J. is supported by the Max Planck Society, Deutsche Forschungsgemeinschaft, Center for Integrated Protein Science Munich, RUBICON EU Network of Excellence, a European Research Council Advanced Grant and the Louis-Jeantet Foundation.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information S1 (figure)
The detection and characterization of DNA–protein crosslinks. (PDF 144 kb)
Rights and permissions
About this article
Cite this article
Stingele, J., Jentsch, S. DNA–protein crosslink repair. Nat Rev Mol Cell Biol 16, 455–460 (2015). https://doi.org/10.1038/nrm4015
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrm4015
This article is cited by
-
Dimerization-dependent serine protease activity of FAM111A prevents replication fork stalling at topoisomerase 1 cleavage complexes
Nature Communications (2024)
-
Comparative transcriptome and metabolome analyses reveal the methanol dissimilation pathway of Pichia pastoris
BMC Genomics (2022)
-
DNA–protein crosslink proteases in genome stability
Communications Biology (2021)
-
FAM111A protects replication forks from protein obstacles via its trypsin-like domain
Nature Communications (2020)
-
Molecular mechanisms of topoisomerase 2 DNA–protein crosslink resolution
Cellular and Molecular Life Sciences (2020)