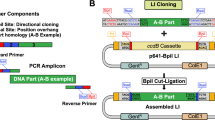
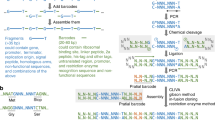
Abstract
DNA assembly is a key part of constructing gene expression systems and even whole chromosomes. In the past decade, a plethora of powerful new DNA assembly methods — including Gibson Assembly, Golden Gate and ligase cycling reaction (LCR) — have been developed. In this Innovation article, we discuss these methods as well as standards such as the modular cloning (MoClo) system, GoldenBraid, modular overlap-directed assembly with linkers (MODAL) and PaperClip, which have been developed to facilitate a streamlined assembly workflow, to aid the exchange of material between research groups and to create modular reusable DNA parts.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Kosuri, S. & Church, G. M. Large-scale de novo DNA synthesis: technologies and applications. Nat. Methods 11, 499–507 (2014).
Gibson, D. G. et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345 (2009).
Keasling, J. D. Synthetic biology for synthetic chemistry. ACS Chem. Biol. 3, 64–76 (2008).
Cameron, D. E., Bashor, C. J. & Collins, J. J. A brief history of synthetic biology. Nat. Rev. Microbiol. 12, 381–390 (2014).
Czar, M. J., Anderson, J. C., Bader, J. S. & Peccoud, J. Gene synthesis demystified. Trends Biotechnol. 27, 63–72 (2009).
Annaluru, N. et al. Total synthesis of a functional designer eukaryotic chromosome. Science 344, 55–58 (2014).
Ellis, T., Adie, T. & Baldwin, G. S. DNA assembly for synthetic biology: from parts to pathways and beyond. Integr. Biol. 3, 109–118 (2011).
Chao, R., Yuan, Y. & Zhao, H. Recent advances in DNA assembly technologies. FEMS Yeast Res. 15, 1–9 (2015).
Casini, A. et al. One-pot DNA construction for synthetic biology: the Modular Overlap-Directed Assembly with Linkers (MODAL) strategy. Nucleic Acids Res. 42, e7 (2014).
Cardinale, S. & Arkin, A. P. Contextualizing context for synthetic biology — identifying causes of failure of synthetic biological systems. Biotechnol. J. 7, 856–866 (2012).
Cohen, S. N., Chang, A. C., Boyer, H. W. & Helling, R. B. Construction of biologically functional bacterial plasmids in vitro. Proc. Natl Acad. Sci. USA 70, 3240–3244 (1973).
Crook, N. C., Freeman, E. S. & Alper, H. S. Re-engineering multicloning sites for function and convenience. Nucleic Acids Res. 39, e92 (2011).
Shetty, R. P., Endy, D. & Knight, T. F. Engineering BioBrick vectors from BioBrick parts. J. Biol. Eng. 2, 5 (2008).
Norville, J. E. et al. Introduction of customized inserts for streamlined assembly and optimization of BioBrick synthetic genetic circuits. J. Biol. Eng. 4, 17 (2010).
Anderson, J. C. et al. BglBricks: a flexible standard for biological part assembly. J. Biol. Eng. 4, 1 (2010).
Leguia, M., Brophy, J. A., Densmore, D., Asante, A. & Anderson, J. C. 2ab assembly: a methodology for automatable, high-throughput assembly of standard biological parts. J. Biol. Eng. 7, 2 (2013).
Litcofsky, K. D., Afeyan, R. B., Krom, R. J., Khalil, A. S. & Collins, J. J. Iterative plug-and-play methodology for constructing and modifying synthetic gene networks. Nat. Methods 9, 1077–1080 (2012).
Silva-Rocha, R. et al. The Standard European Vector Architecture (SEVA): a coherent platform for the analysis and deployment of complex prokaryotic phenotypes. Nucleic Acids Res. 41, D666–D675 (2013).
Martínez-García, E., Aparicio, T., Goñi-Moreno, A., Fraile, S. & de Lorenzo, V. SEVA 2.0: an update of the Standard European Vector Architecture for de-/re-construction of bacterial functionalities. Nucleic Acids Res. 43, D1183–D1189 (2014).
Marcaida, M. J., Muñoz, I. G., Blanco, F. J., Prieto, J. & Montoya, G. Homing endonucleases: from basics to therapeutic applications. Cell. Mol. Life Sci. 67, 727–748 (2010).
Liu, J.-K., Chen, W.-H., Ren, S.-X., Zhao, G.-P. & Wang, J. iBrick: a new standard for iterative assembly of biological parts with homing endonucleases. PLoS ONE 9, e110852 (2014).
Li, M. V. et al. HomeRun vector assembly system: a flexible and standardized cloning system for assembly of multi-modular DNA constructs. PLoS ONE 9, e100948 (2014).
Sleight, S. C. & Sauro, H. M. Visualization of evolutionary stability dynamics and competitive fitness of Escherichia coli engineered with randomized multi-gene circuits. ACS Synth. Biol. 2, 519–528 (2013).
Engler, C., Kandzia, R. & Marillonnet, S. A one pot, one step, precision cloning method with high throughput capability. PLoS ONE 3, e3647 (2008).
Cermak, T. et al. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 39, e82 (2011).
Kamens, J. The Addgene repository: an international nonprofit plasmid and data resource. Nucleic Acids Res. 43, D1152–D1157 (2015).
Mutalik, V. K. et al. Precise and reliable gene expression via standard transcription and translation initiation elements. Nat. Methods 10, 354–360 (2013).
Linshiz, G. et al. PR-PR: cross-platform laboratory automation system. ACS Synth. Biol. 3, 515–524 (2014).
Weber, E., Engler, C., Gruetzner, R., Werner, S. & Marillonnet, S. A modular cloning system for standardized assembly of multigene constructs. PLoS ONE 6, e16765 (2011).
Sarrion-Perdigones, A. et al. GoldenBraid 2.0: a comprehensive DNA assembly framework for plant synthetic biology. Plant Physiol. 162, 1618–1631 (2013).
Binder, A. et al. A modular plasmid assembly kit for multigene expression, gene silencing and silencing rescue in plants. PLoS ONE 9, e88218 (2014).
Lampropoulos, A. et al. GreenGate — a novel, versatile, and efficient cloning system for plant transgenesis. PLoS ONE 8, e83043 (2013).
Sarrion-Perdigones, A. et al. GoldenBraid: an iterative cloning system for standardized assembly of reusable genetic modules. PLoS ONE 6, e21622 (2011).
Duportet, X. et al. A platform for rapid prototyping of synthetic gene networks in mammalian cells. Nucleic Acids Res. 42, 13440–13451 (2014).
Sakuma, T., Nishikawa, A., Kume, S., Chayama, K. & Yamamoto, T. Multiplex genome engineering in human cells using all-in-one CRISPR/Cas9 vector system. Sci. Rep. 4, 5400 (2014).
Chen, W.-H., Qin, Z.-J., Wang, J. & Zhao, G.-P. The MASTER (methylation-assisted tailorable ends rational) ligation method for seamless DNA assembly. Nucleic Acids Res. 41, e93 (2013).
Blake, W. J. et al. Pairwise selection assembly for sequence-independent construction of long-length DNA. Nucleic Acids Res. 38, 2594–2602 (2010).
Hartley, J. L., Temple, G. F. & Brasch, M. A. DNA cloning using in vitro site-specific recombination. Genome Res. 10, 1788–1795 (2000).
Alberti, S., Gitler, A. D. & Lindquist, S. A suite of Gateway cloning vectors for high-throughput genetic analysis in Saccharomyces cerevisiae. Yeast 24, 913–919 (2007).
Marsischky, G. & LaBaer, J. Many paths to many clones: a comparative look at high-throughput cloning methods. Genome Res. 14, 2020–2028 (2004).
Sasaki, Y. et al. Evidence for high specificity and efficiency of multiple recombination signals in mixed DNA cloning by the multisite Gateway system. J. Biotechnol. 107, 233–243 (2004).
Zhang, L., Zhao, G. & Ding, X. Tandem assembly of the epothilone biosynthetic gene cluster by in vitro site-specific recombination. Sci. Rep. 1, 141 (2011).
Colloms, S. D. et al. Rapid metabolic pathway assembly and modification using serine integrase site-specific recombination. Nucleic Acids Res. 42, e23 (2013).
Horton, R. M., Hunt, H. D., Ho, S. N., Pullen, J. K. & Pease, L. R. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77, 61–68 (1989).
Quan, J. & Tian, J. Circular polymerase extension cloning of complex gene libraries and pathways. PLoS ONE 4, e6441 (2009).
Bitinaite, J. et al. USER friendly DNA engineering and cloning method by uracil excision. Nucleic Acids Res. 35, 1992–2002 (2007).
Annaluru, N. et al. Assembling DNA fragments by USER fusion. Methods Mol. Biol. 852, 77–95 (2012).
Li, M. Z. & Elledge, S. J. Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nat. Methods 4, 251–256 (2007).
Zhang, Y., Werling, U. & Edelmann, W. SLiCE: a novel bacterial cell extract-based DNA cloning method. Nucleic Acids Res. 40, e55 (2012).
Itaya, M., Fujita, K., Kuroki, A. & Tsuge, K. Bottom-up genome assembly using the Bacillus subtilis genome vector. Nat. Methods 5, 41–43 (2008).
Raymond, C. K., Pownder, T. A. & Sexson, S. L. General method for plasmid construction using homologous recombination. Biotechniques 26, 134–141 (1999).
Gibson, D. G. et al. One-step assembly in yeast of 25 overlapping DNA fragments to form a complete synthetic Mycoplasma genitalium genome. Proc. Natl Acad. Sci. USA 105, 20404–20409 (2008).
Shao, Z., Zhao, H. & Zhao, H. DNA assembler, an in vivo genetic method for rapid construction of biochemical pathways. Nucleic Acids Res. 37, e16 (2009).
Casini, A. et al. R2oDNA designer: computational design of biologically neutral synthetic DNA sequences. ACS Synth. Biol. 3, 525–528 (2014).
Torella, J. P. et al. Rapid construction of insulated genetic circuits via synthetic sequence-guided isothermal assembly. Nucleic Acids Res. 42, 681–689 (2013).
Trubitsyna, M., Michlewski, G., Cai, Y., Elfick, A. & French, C. E. PaperClip: rapid multi-part DNA assembly from existing libraries. Nucleic Acids Res. 42, e154 (2014).
de Kok, S. et al. Rapid and reliable DNA assembly via ligase cycling reaction. ACS Synth. Biol. 3, 97–106 (2014).
Storch, M. et al. BASIC: a new biopart assembly standard provides accurate, single-tier DNA assembly for synthetic biology. ACS Synth. Biol. http://dx.doi.org/10.1021/sb500356d (2015).
Guye, P., Li, Y., Wroblewska, L., Duportet, X. & Weiss, R. Rapid, modular and reliable construction of complex mammalian gene circuits. Nucleic Acids Res. 41, e156 (2013).
Kahl, L. J. & Endy, D. A survey of enabling technologies in synthetic biology. J. Biol. Eng. 7, 13 (2013).
Gibson, D. G., Smith, H. O., Hutchison, C. A., Venter, J. C. & Merryman, C. Chemical synthesis of the mouse mitochondrial genome. Nat. Methods 7, 901–903 (2010).
Smanski, M. J. et al. Functional optimization of gene clusters by combinatorial design and assembly. Nat. Biotechnol. 32, 1241–1249 (2014).
Gibson, D. G. et al. Complete chemical synthesis, assembly, and cloning of a Mycoplasma genitalium genome. Science 319, 1215–1220 (2008).
Gibson, D. G. et al. Creation of a bacterial cell controlled by a chemically synthesized genome. Science 329, 52–56 (2010).
Jovicevic, D., Blount, B. A. & Ellis, T. Total synthesis of a eukaryotic chromosome: redesigning and SCRaMbLE-ing yeast. Bioessays 36, 855–860 (2014).
Dymond, J. S. et al. Synthetic chromosome arms function in yeast and generate phenotypic diversity by design. Nature 477, 471–476 (2011).
Hillson, N. J., Rosengarten, R. D. & Keasling, J. D. j5 DNA assembly design automation software. ACS Synth. Biol. 1, 14–21 (2012).
Appleton, E., Tao, J., Haddock, T. & Densmore, D. Interactive assembly algorithms for molecular cloning. Nat. Methods 11, 657–662 (2014).
Densmore, D. et al. Algorithms for automated DNA assembly. Nucleic Acids Res. 38, 2607–2616 (2010).
Wang, H. H. et al. Programming cells by multiplex genome engineering and accelerated evolution. Nature 460, 894–898 (2009).
Lajoie, M. J. et al. Genomically recoded organisms expand biological functions. Science 342, 357–360 (2013).
Datsenko, K. A. & Wanner, B. L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA 97, 6640–6645 (2000).
Pósfai, G. et al. Emergent properties of reduced-genome Escherichia coli. Science 312, 1044–1046 (2006).
Kim, H. & Kim, J.-S. A guide to genome engineering with programmable nucleases. Nat. Rev. Genet. 15, 321–334 (2014).
Horwitz, A. A. et al. Efficient multiplexed integration of synergistic alleles and metabolic pathways in yeasts via CRISPR–Cas. Cell Syst. http://dx.doi.org/10.1016/j.cels.2015.02.001 (2015).
Groth, A. C. & Calos, M. P. Phage integrases: biology and applications. J. Mol. Biol. 335, 667–678 (2004).
Torella, J. P. et al. Unique nucleotide sequence-guided assembly of repetitive DNA parts for synthetic biology applications. Nat. Protoc. 9, 2075–2089 (2014).
Acknowledgements
The authors thank M. Jamilly for contributing to initial discussions and apologize to those whose work was not discussed owing to space limitations. Research on DNA assembly in the groups of T.E. and G.S.B. is supported by the UK Engineering and Physical Research Council (EPSRC) grant EP/J02175X/1 and EU FP7 grant KBBE.2011.5-289326. M.S. is supported by Marie Curie Intra-European Fellowship 628019.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Casini, A., Storch, M., Baldwin, G. et al. Bricks and blueprints: methods and standards for DNA assembly. Nat Rev Mol Cell Biol 16, 568–576 (2015). https://doi.org/10.1038/nrm4014
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrm4014
This article is cited by
-
Current achievements, strategies, obstacles, and overcoming the challenges of the protein engineering in Pichia pastoris expression system
World Journal of Microbiology and Biotechnology (2024)
-
DNA synthesis technologies to close the gene writing gap
Nature Reviews Chemistry (2023)
-
A framework to efficiently describe and share reproducible DNA materials and construction protocols
Nature Communications (2022)
-
Epigenetic silencing directs expression heterogeneity of stably integrated multi-transcript unit genetic circuits
Scientific Reports (2021)
-
Single strain control of microbial consortia
Nature Communications (2021)