Key Points
-
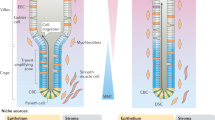
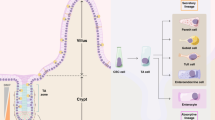
Intestinal stem cells are responsible for the remarkable ability of the intestinal epithelium to efficiently renew and repair itself throughout life.
-
The intestinal stem cells reside within specialized instructive niches at the crypt base. WNT is an essential niche factor supplied by both Paneth cells and non-epithelial cells.
-
The identification of new intestinal stem cell markers has greatly improved our understanding of stem cell biology during homeostasis and disease.
-
Intestinal stem cells can now be purified and used to grow new epithelia ex vivo for regenerative medicine applications.
-
'Reserve' stem cells are quickly recruited to drive epithelial regeneration following loss of the regular stem cell pool due to injury.
Abstract
Small populations of adult stem cells are responsible for the remarkable ability of the epithelial lining of the intestine to be efficiently renewed and repaired throughout life. The recent discovery of specific markers for these stem cells, together with the development of new technologies to track endogenous stem cell activity in vivo and to exploit their ability to generate new epithelia ex vivo, has greatly improved our understanding of stem cell-driven homeostasis, regeneration and cancer in the intestine. These exciting new insights into the biology of intestinal stem cells have the potential to accelerate the development of stem cell-based therapies and ameliorate cancer treatments.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Leblond, C. P. & Walker, B. E. Renewal of cell populations. Physiol. Rev. 36, 255–276 (1956).
Barker, N., van de Wetering, M. & Clevers, H. The intestinal stem cell. Genes Dev. 22, 1856–1864 (2008).
Potten, C. S., Gandara, R., Mahida, Y. R., Loeffler, M. & Wright, N. A. The stem cells of small intestinal crypts: where are they? Cell Prolif. 42, 731–750 (2009).
Blanpain, C. & Simons, B. D. Unravelling stem cell dynamics by lineage tracing. Nature Rev. Mol. Cell Biol. 14, 489–502 (2013).
Sato, T. & Clevers, H. Growing self-organizing mini-guts from a single intestinal stem cell: mechanism and applications. Science 340, 1190–1194 (2013).
Barker, N., Bartfeld, S. & Clevers, H. Tissue-resident adult stem cell populations of rapidly self-renewing organs. Cell Stem Cell 7, 656–670 (2010).
Buczacki, S. J. et al. Intestinal label-retaining cells are secretory precursors expressing Lgr5. Nature 495, 65–69 (2013). Identified LRCs as Paneth cell progenitors capable of reverting to stem cells after damage.
Tian, H. et al. A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature 478, 255–259 (2011). Demonstrated that the intestinal epithelium is able to survive ablation of the LGR5+ stem cell compartment, indicating the existence of reserve stem cells.
Yilmaz, O. H. et al. mTORC1 in the Paneth cell niche couples intestinal stem-cell function to calorie intake. Nature 486, 490–495 (2012).
Gerbe, F., Legraverend, C. & Jay, P. The intestinal epithelium tuft cells: specification and function. Cell. Mol. Life Sci. 69, 2907–2917 (2012).
Watson, A. J. & Hughes, K. R. TNF-α-induced intestinal epithelial cell shedding: implications for intestinal barrier function. Ann. NY Acad. Sci. 1258, 1–8 (2012).
Bjerknes, M. & Cheng, H. Gastrointestinal stem cells. II. Intestinal stem cells. Am. J. Physiol. Gastrointest. Liver Physiol. 289, G381–G387 (2005).
Ireland, H., Houghton, C., Howard, L. & Winton, D. J. Cellular inheritance of a Cre-activated reporter gene to determine Paneth cell longevity in the murine small intestine. Dev. Dyn. 233, 1332–1336 (2005).
Grosse, A. S. et al. Cell dynamics in fetal intestinal epithelium: implications for intestinal growth and morphogenesis. Development 138, 4423–4432 (2011).
Gregorieff, A. & Clevers, H. Wnt signaling in the intestinal epithelium: from endoderm to cancer. Genes Dev. 19, 877–890 (2005).
Madison, B. B. et al. Epithelial hedgehog signals pattern the intestinal crypt-villus axis. Development 132, 279–289 (2005).
Korinek, V. et al. Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nature Genet. 19, 379–383 (1998).
Cheng, H. & Bjerknes, M. Whole population cell kinetics and postnatal development of the mouse intestinal epithelium. Anat. Rec. 211, 420–426 (1985).
Dehmer, J. J. et al. Expansion of intestinal epithelial stem cells during murine development. PLoS ONE 6, e27070 (2011).
Mustata, R. C. et al. Identification of Lgr5-independent spheroid-generating progenitors of the mouse fetal intestinal epithelium. Cell Rep. 5, 421–432 (2013).
Itzkovitz, S., Blat, I. C., Jacks, T., Clevers, H. & van Oudenaarden, A. Optimality in the development of intestinal crypts. Cell 148, 608–619 (2012).
Fordham, R. P. et al. Transplantation of expanded fetal intestinal progenitors contributes to colon regeneration after injury. Cell Stem Cell http://dx.doi.org/10.1016/j.stem.2013.09.015 (2013).
Ponder, B. A., Festing, M. F. & Wilkinson, M. M. An allelic difference determines reciprocal patterns of expression of binding sites for Dolichos biflorus lectin in inbred strains of mice. J. Embryol. Exp. Morphol. 87, 229–239 (1985).
Schmidt, G. H., Garbutt, D. J., Wilkinson, M. M. & Ponder, B. A. Clonal analysis of intestinal crypt populations in mouse aggregation chimaeras. J. Embryol. Exp. Morphol. 85, 121–130 (1985).
Ponder, B. A. et al. Derivation of mouse intestinal crypts from single progenitor cells. Nature 313, 689–691 (1985). Showed that adult crypts harbour clonal stem cell populations derived from a single progenitor cell.
Hermiston, M. L., Green, R. P. & Gordon, J. I. Chimeric-transgenic mice represent a powerful tool for studying how the proliferation and differentiation programs of intestinal epithelial cell lineages are regulated. Proc. Natl Acad. Sci. USA 90, 8866–8870 (1993).
Bjerknes, M. & Cheng, H. Clonal analysis of mouse intestinal epithelial progenitors. Gastroenterology 116, 7–14 (1999).
Gutierrez-Gonzalez, L. et al. Analysis of the clonal architecture of the human small intestinal epithelium establishes a common stem cell for all lineages and reveals a mechanism for the fixation and spread of mutations. J. Pathol. 217, 489–496 (2009).
Greaves, L. C. et al. Mitochondrial DNA mutations are established in human colonic stem cells, and mutated clones expand by crypt fission. Proc. Natl Acad. Sci. USA 103, 714–719 (2006).
Taylor, R. W. et al. Mitochondrial DNA mutations in human colonic crypt stem cells. J. Clin. Invest. 112, 1351–1360 (2003). Established the clonality of human crypt stem cell populations by examining mitochondrial mutation patterns.
Novelli, M. R. et al. Polyclonal origin of colonic adenomas in an XO/XY patient with FAP. Science 272, 1187–1190 (1996). Used chimeric human patient cells to show that human colonic crypts are maintained by a clonal population of adult stem cells.
Hendry, J. H., Roberts, S. A. & Potten, C. S. The clonogen content of murine intestinal crypts: dependence on radiation dose used in its determination. Radiat. Res. 132, 115–119 (1992).
Potten, C. S. & Loeffler, M. A comprehensive model of the crypts of the small intestine of the mouse provides insight into the mechanisms of cell migration and the proliferation hierarchy. J. Theor. Biol. 127, 381–391 (1987).
Bjerknes, M. & Cheng, H. The stem-cell zone of the small intestinal epithelium. III. Evidence from columnar, enteroendocrine, and mucous cells in the adult mouse. Am. J. Anat. 160, 77–91 (1981).
Kozar, S. et al. Continuous clonal labeling reveals small numbers of functional stem cells in intestinal crypts and adenomas. Cell Stem Cell 13, 626–633 (2013).
Winton, D. J., Blount, M. A. & Ponder, B. A. A clonal marker induced by mutation in mouse intestinal epithelium. Nature 333, 463–466 (1988). Reported the existence of multipotent, self-renewing stem cells in the adult small intestine, as shown by the use of a random somatic mutation approach.
Cairnie, A. B., Lamerton, L. F. & Steel, G. G. Cell proliferation studies in the intestinal epithelium of the rat. I. Determination of the kinetic parameters. Exp. Cell Res. 39, 528–538 (1965).
Qiu, J. M., Roberts, S. A. & Potten, C. S. Cell migration in the small and large bowel shows a strong circadian rhythm. Epithelial Cell Biol. 3, 137–148 (1994). Together with reference 37, this paper examined cell migration rates in the intestinal epithelium and proposed position +4 as the origin of cell migration.
Cairns, J. Mutation selection and natural history of cancer. Nature 255, 197–200 (1975).
Potten, C. S., Booth, C. & Pritchard, D. M. The intestinal epithelial stem cell: the mucosal governor. Int. J. Exp. Pathol. 78, 219–243 (1997).
Cheng, H. & Leblond, C. P. Origin, differentiation and renewal of the four main epithelial cell types in the mouse small intestine. V. Unitarian Theory of the origin of the four epithelial cell types. Am. J. Anat. 141, 537–561 (1974). Part of a seminal series of papers that described a probable common CBC cell origin for the four main epithelial cell lineages.
Cheng, H. & Leblond, C. P. Origin, differentiation and renewal of the four main epithelial cell types in the mouse small intestine. I. Columnar cell. Am. J. Anat. 141, 461–479 (1974).
Cheng, H. & Leblond, C. P. Origin, differentiation and renewal of the four main epithelial cell types in the mouse small intestine. III. Entero-endocrine cells. Am. J. Anat. 141, 503–519 (1974).
Cheng, H. Origin, differentiation and renewal of the four main epithelial cell types in the mouse small intestine. IV. Paneth cells. Am. J. Anat. 141, 521–535 (1974).
Cheng, H. Origin, differentiation and renewal of the four main epithelial cell types in the mouse small intestine. II. Mucous cells. Am. J. Anat. 141, 481–501 (1974).
Barker, N. et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449, 1003–1007 (2007). Reported the validation of LGR5 as a marker of CBC stem cells in the small intestine and colon using in vivo lineage tracing.
Sato, T. et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–265 (2009). Described a novel ex vivo culture system capable of sustaining the long-term growth of near-physiological intestinal epithelia from isolated LGR5+ stem cells.
Potten, C. S. Extreme sensitivity of some intestinal crypt cells to X and y-irradiation. Nature 269, 518–521 (1977). Analysed the sensitivity of epithelial populations at various locations within the crypt to irradiation, showing that +4 cells are highly radiosensitive.
Lansdorp, P. M. Immortal strands? Give me a break. Cell 129, 1244–1247 (2007).
Jung, P. et al. Isolation and in vitro expansion of human colonic stem cells. Nature Med. 17, 1225–1227 (2011).
Munoz, J. et al. The Lgr5 intestinal stem cell signature: robust expression of proposed quiescent '+4' cell markers. EMBO J. 31, 3079–3091 (2012).
Van der Flier, L. G. et al. The intestinal Wnt/TCF signature. Gastroenterology 132, 628–632 (2007).
van der Flier, L. G. et al. Transcription factor achaete scute-like 2 controls intestinal stem cell fate. Cell 136, 903–912 (2009).
de Lau, W. et al. Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature 476, 293–297 (2011).
Carmon, K. S., Gong, X., Lin, Q., Thomas, A. & Liu, Q. R-spondins function as ligands of the orphan receptors LGR4 and LGR5 to regulate Wnt/β-catenin signaling. Proc. Natl Acad. Sci. USA 108, 11452–11457 (2011).
Carmon, K. S., Lin, Q., Gong, X., Thomas, A. & Liu, Q. LGR5 interacts and co-internalizes with Wnt receptors to modulate Wnt/β-catenin signaling. Mol Cell. Biol. 32, 2054–2064 http://dx.doi.org/10.1128/MCB.00272-12 (2012).
Glinka, A. et al. LGR4 and LGR5 are R-spondin receptors mediating Wnt/β-catenin and Wnt/PCP signalling. EMBO Rep. 12, 1055–1061 (2011).
Koo, B.-K. et al. Tumour suppressor RNF43 is a stem-cell E3 ligase that induces endocytosis of Wnt receptors. Nature 488, 665–669 (2012).
Hao, H. X. et al. ZNRF3 promotes Wnt receptor turnover in an R-spondin-sensitive manner. Nature 485, 195–200 (2012).
Fafilek, B. et al. Troy, a tumor necrosis factor receptor family member, interacts with lgr5 to inhibit wnt signaling in intestinal stem cells. Gastroenterology 144, 381–391 (2013).
Chen, P. H., Chen, X., Lin, Z., Fang, D. & He, X. The structural basis of R-spondin recognition by LGR5 and RNF43. Genes Dev. 27, 1345–1350 (2013).
Peng, W. C. et al. Structure of stem cell growth factor R-spondin 1 in complex with the ectodomain of its receptor LGR5. Cell Rep. 3, 1885–1892 (2013).
Wang, D. et al. Structural basis for R-spondin recognition by LGR4/5/6 receptors. Genes Dev. 27, 1339–1344 (2013).
Snippert, H. J. et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell 143, 134–144 (2010).
Lopez-Garcia, C., Klein, A. M., Simons, B. D. & Winton, D. J. Intestinal stem cell replacement follows a pattern of neutral drift. Science 330, 822–825 (2010). References 64 and 65 report the use of multicolor tracing and computer modelling to examine the population dynamics of Lgr5+ stem cells in the small intestine
Quyn, A. J. et al. Spindle orientation bias in gut epithelial stem cell compartments is lost in precancerous tissue. Cell Stem Cell 6, 175–181 (2010).
Bellis, J. et al. The tumor suppressor Apc controls planar cell polarities central to gut homeostasis. J. Cell Biol. 198, 331–341 (2012).
Escobar, M. et al. Intestinal epithelial stem cells do not protect their genome by asymmetric chromosome segregation. Nature Commun. 2, 258 (2011).
Schepers, A. G., Vries, R., van den Born, M., van de Wetering, M. & Clevers, H. Lgr5 intestinal stem cells have high telomerase activity and randomly segregate their chromosomes. EMBO J. 30, 1104–1109 (2011).
Steinhauser, M. L. et al. Multi-isotope imaging mass spectrometry quantifies stem cell division and metabolism. Nature 481, 516–519 (2012).
Buske, P. et al. On the biomechanics of stem cell niche formation in the gut — modelling growing organoids. FEBS J. 279, 3475–3487 (2012).
Buske, P. et al. A comprehensive model of the spatio-temporal stem cell and tissue organisation in the intestinal crypt. PLoS Comput. Biol. 7, e1001045 (2011).
Kaaij, L. T. et al. DNA methylation dynamics during intestinal stem cell differentiation reveals enhancers driving gene expression in the villus. Genome Biol. 14, R50 (2013).
Sakamori, R. et al. Cdc42 and Rab8a are critical for intestinal stem cell division, survival, and differentiation in mice. J. Clin. Invest. 122, 1052–1065 (2012).
Heijmans, J. et al. ER stress causes rapid loss of intestinal epithelial stemness through activation of the unfolded protein response. Cell Rep. 3, 1128–1139 (2013).
Sangiorgi, E. & Capecchi, M. R. Bmi1 is expressed in vivo in intestinal stem cells. Nature Genet. 40, 915–920 (2008). Reported the first validation of a +4 marker by in vivo lineage tracing.
Yan, K. S. et al. The intestinal stem cell markers Bmi1 and Lgr5 identify two functionally distinct populations. Proc. Natl Acad. Sci. USA 109, 466–471 (2012).
Itzkovitz, S. et al. Single-molecule transcript counting of stem-cell markers in the mouse intestine. Nature Cell Biol. 14, 106–114 (2012).
Montgomery, R. K. et al. Mouse telomerase reverse transcriptase (mTert) expression marks slowly cycling intestinal stem cells. Proc. Natl Acad. Sci. USA 108, 179–184 (2011).
Powell, A. E. et al. The pan-ErbB negative regulator Lrig1 is an intestinal stem cell marker that functions as a tumor suppressor. Cell 149, 146–158 (2012).
Takeda, N. et al. Interconversion between intestinal stem cell populations in distinct niches. Science 334, 1420–1424 (2011). Showed that +4 stem cells can interconvert with LGR5+ stem cells.
Jensen, K. B. et al. Lrig1 expression defines a distinct multipotent stem cell population in mammalian epidermis. Cell Stem Cell 4, 427–439 (2009).
Wong, V. W. et al. Lrig1 controls intestinal stem-cell homeostasis by negative regulation of ErbB signalling. Nature Cell Biol. 14, 401–408 (2012).
Breault, D. T. et al. Generation of mTert–GFP mice as a model to identify and study tissue progenitor cells. Proc. Natl Acad. Sci. USA 105, 10420–10425 (2008).
Roberts, S. A., Hendry, J. H. & Potten, C. S. Deduction of the clonogen content of intestinal crypts: a direct comparison of two-dose and multiple-dose methodologies. Radiat. Res. 141, 303–308 (1995).
Sato, T. et al. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature 469, 415–418 (2011).
Roth, S. et al. Paneth cells in intestinal homeostasis and tissue injury. PLoS ONE 7, e38965 (2012).
van Es, J. H. et al. Dll1+ secretory progenitor cells revert to stem cells upon crypt damage. Nature Cell Biol. 14, 1099–1104 (2012). Showed that DLL1+ secretory progenitors are the reserve stem cell population activated in response to injury.
Kemper, K. et al. Monoclonal antibodies against Lgr5 identify human colorectal cancer stem cells. Stem Cells 30, 2378–2386 (2012).
Garabedian, E. M., Roberts, L. J., McNevin, M. S. & Gordon, J. I. Examining the role of Paneth cells in the small intestine by lineage ablation in transgenic mice. J. Biol. Chem. 272, 23729–23740 (1997).
Durand, A. et al. Functional intestinal stem cells after Paneth cell ablation induced by the loss of transcription factor Math1 (Atoh1). Proc. Natl Acad. Sci. USA 109, 8965–8970 (2012).
Kim, T. H., Escudero, S. & Shivdasani, R. A. Intact function of Lgr5 receptor-expressing intestinal stem cells in the absence of Paneth cells. Proc. Natl Acad. Sci. USA 109, 3932–3937 (2012).
Farin, H. F., Van Es, J. H. & Clevers, H. Redundant sources of Wnt regulate intestinal stem cells and promote formation of Paneth cells. Gastroenterology 143, 1518–1529.e7 (2012).
Rothenberg, M. E. et al. Identification of a cKit+ colonic crypt base secretory cell that supports Lgr5+ stem cells in mice. Gastroenterology 142, 1195–1205.e6 (2012).
Blache, P. et al. SOX9 is an intestine crypt transcription factor, is regulated by the Wnt pathway, and represses the CDX2 and MUC2 genes. J. Cell Biol. 166, 37–47 (2004).
Bastide, P. et al. Sox9 regulates cell proliferation and is required for Paneth cell differentiation in the intestinal epithelium. J. Cell Biol. 178, 635–648 (2007).
Formeister, E. J. et al. Distinct SOX9 levels differentially mark stem/progenitor populations and enteroendocrine cells of the small intestine epithelium. Am. J. Physiol. Gastrointest. Liver Physiol. 296, G1108–G1118 (2009).
Van Landeghem, L. et al. Activation of two distinct Sox9-EGFP-expressing intestinal stem cell populations during crypt regeneration after irradiation. Am. J. Physiol. Gastrointest. Liver Physiol. 302, G1111–G1132 (2012).
Furuyama, K. et al. Continuous cell supply from a Sox9-expressing progenitor zone in adult liver, exocrine pancreas and intestine. Nature Genet. 43, 34–41 (2011).
Gracz, A. D., Ramalingam, S. & Magness, S. T. Sox9 expression marks a subset of CD24-expressing small intestine epithelial stem cells that form organoids in vitro. Am. J. Physiol. Gastrointest Liver Physiol. 298, G590–G600 (2010).
Potten, C. S. et al. Identification of a putative intestinal stem cell and early lineage marker; musashi-1. Differentiation 71, 28–41 (2003).
Kayahara, T. et al. Candidate markers for stem and early progenitor cells, Musashi-1 and Hes1, are expressed in crypt base columnar cells of mouse small intestine. FEBS Lett. 535, 131–135 (2003).
Cambuli, F. M., Rezza, A., Nadjar, J. & Plateroti, M. Musashi1-eGFP mice, a new tool for differential isolation of the intestinal stem cell populations. Stem Cells 31, 2273–2278 (2013).
Takeda, H., Koso, H., Tessarollo, L., Copeland, N. G. & Jenkins, N. A. Musashi1-CreERT2: a new cre line for conditional mutagenesis in neural stem cells. Genesis 51, 128–134 (2013).
O'Brien, C. A., Pollett, A., Gallinger, S. & Dick, J. E. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature 445, 106–110 (2007).
Shmelkov, S. V. et al. CD133 expression is not restricted to stem cells, and both CD133+ and CD133− metastatic colon cancer cells initiate tumors. J. Clin. Invest. 118, 2111–2120 (2008).
Yin, A. H. et al. AC133, a novel marker for human hematopoietic stem and progenitor cells. Blood 90, 5002–5012 (1997).
Zhu, L. et al. Prominin 1 marks intestinal stem cells that are susceptible to neoplastic transformation. Nature 457, 603–607 (2009).
Snippert, H. J. et al. Prominin-1/CD133 marks stem cells and early progenitors in mouse small intestine. Gastroenterology 136, 2187–2194 (2009).
Barker, N., van Oudenaarden, A. & Clevers, H. Identifying the stem cell of the intestinal crypt: strategies and pitfalls. Cell Stem Cell 11, 452–460 (2012).
He, X. C. et al. PTEN-deficient intestinal stem cells initiate intestinal polyposis. Nature Genet. 39, 189–198 (2007).
He, X. C. et al. BMP signaling inhibits intestinal stem cell self-renewal through suppression of Wnt-β-catenin signaling. Nature Genet. 36, 1117–1121 (2004).
Bjerknes, M. & Cheng, H. Re-examination of P-PTEN staining patterns in the intestinal crypt. Nature Genet. 37, 1016–1017; author reply 1017–1018 (2005).
Demidov, O. N. et al. Wip1 phosphatase regulates p53-dependent apoptosis of stem cells and tumorigenesis in the mouse intestine. Cell Stem Cell 1, 180–190 (2007).
Giannakis, M. et al. Molecular properties of adult mouse gastric and intestinal epithelial progenitors in their niches. J. Biol. Chem. 281, 11292–11300 (2006).
May, R. et al. Identification of a novel putative gastrointestinal stem cell and adenoma stem cell marker, doublecortin and CaM kinase-like-1, following radiation injury and in adenomatous polyposis coli/multiple intestinal neoplasia mice. Stem Cells 26, 630–637 (2008).
May, R. et al. Doublecortin and CaM kinase-like-1 and leucine-rich-repeat-containing G-protein-coupled receptor mark quiescent and cycling intestinal stem cells, respectively. Stem Cells 27, 2571–2579 (2009).
Gerbe, F., Brulin, B., Makrini, L., Legraverend, C. & Jay, P. DCAMKL-1 expression identifies Tuft cells rather than stem cells in the adult mouse intestinal epithelium. Gastroenterology 137, 2179–2180; author reply 2180–2181 (2009).
Nakanishi, Y. et al. Dclk1 distinguishes between tumor and normal stem cells in the intestine. Nature Genet. 45, 98–103 (2013).
Sato, T. et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett's epithelium. Gastroenterology 141, 1762–1772 (2011).
Sala, F. G. et al. A multicellular approach forms a significant amount of tissue-engineered small intestine in the mouse. Tissue Eng. Part A 17, 1841–1850 (2011).
Yui, S. et al. Functional engraftment of colon epithelium expanded in vitro from a single adult Lgr5+ stem cell. Nature Med. 18, 618–623 (2012). First demonstration that intestinal epithelia cultured ex vivo can be used to repair damaged colonic epithelia in live mice.
Magney, J. E., Erlandsen, S. L., Bjerknes, M. L. & Cheng, H. Morphology of the basal surface and evidence for paracrinelike cells. 177, 43–53 (1986).
Acknowledgements
The author thanks the members of the Barker group for critical input. N.B. is supported by the Agency for Science, Technology and Research (A*STAR).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The author declares no competing financial interests.
Supplementary information
Supplementary information S1 (box)
Stem cells in intestinal cancer (PDF 128 kb)
Glossary
- Niches
-
Specialized instructive microenvironments in which stem cells reside. Niches provide all of the factors necessary to regulate stem cell survival and function.
- Intestinal villus
-
A finger-like structure covered in simple columnar epithelium that projects into the intestinal lumen to maximize the surface area for digestion and absorption.
- Duodenum
-
The proximal third of the small intestine, closest to the stomach. Characterized by the presence of long villi to ensure maximal nutrient digestion and absorption.
- Intestinal crypt
-
Tubular invaginations of the epithelium harbouring the stem cells and their proliferating progeny; responsible for driving epithelial homeostasis and regeneration.
- Chimeric mice
-
Mice that are comprised of two or more populations of genetically distinct cells.
- Lgr5–EGFP–ires–CreERT2/R26R–lacZ mouse model
-
Generated by crossing Lgr5–EGFP–ires–CreERT2 (which encodes a tamoxifen-activatable Cre enzyme that catalyzes recombination across DNA sequences called loxP sites) and R26R (Rosa26 reporter construct)–lacZ mouse strains. Facilitates the conditional activation of the lacZ reporter gene in LGR5-expressing cells in living tissues to evaluate their stem cell identity via lineage tracing.
- Bmi1–ires–CreERT2/R26R–lacZ mouse model
-
Generated by crossing Bmi1-ires-CreERT2 and R26R-lacZ mouse strains. Facilitates the conditional activation of the lacZ reporter gene in BMI1-expressing cells in living tissues to evaluate their stem cell identity via lineage tracing.
- Ires
-
(Internal ribosome entry site). A ribosome-binding site present in the middle of an mRNA that facilitates internal translation initiation to generate an independent protein.
- Pulse–chase
-
A method for detecting quiescent label-retaining cells (LRCs) in vivo. A nucleotide analogue (the label) is administered to the mouse for a short period (the pulse); this is followed by an extended period when no further nucleotide analogue is given (the chase). Actively dividing cells rapidly dilute out the label during DNA replication, and labelled cells are lost within 3–4 rounds of cell division. Non-dividing (quiescent) cells do not dilute out the label and thus remain detectable as LRCs.
Rights and permissions
About this article
Cite this article
Barker, N. Adult intestinal stem cells: critical drivers of epithelial homeostasis and regeneration. Nat Rev Mol Cell Biol 15, 19–33 (2014). https://doi.org/10.1038/nrm3721
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrm3721
This article is cited by
-
Intestinal stem cells: guardians of homeostasis in health and aging amid environmental challenges
Experimental & Molecular Medicine (2024)
-
Paracrine signalling by pancreatic δ cells determines the glycaemic set point in mice
Nature Metabolism (2024)
-
Loss of Mptx2 alters bacteria composition and intestinal homeostasis potentially by impairing autophagy
Communications Biology (2024)
-
Pioneering gut health improvements in piglets with phytogenic feed additives
Applied Microbiology and Biotechnology (2024)
-
Human milk oligosaccharides promote intestinal epithelium regeneration independent of the microbiota during necrotizing enterocolitis
Pediatric Surgery International (2024)