Key Points
-
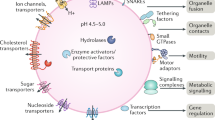
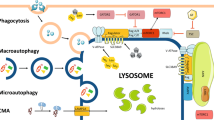
Lysosomes are cellular organelles involved in the degradation and recycling of cellular waste. Extracellular and intracellular materials to be degraded reach the lysosome via endocytosis and autophagy, respectively. Lysosomes are also involved in secretion and plasma membrane repair, by fusing to the plasma membrane in a process termed lysosomal exocytosis.
-
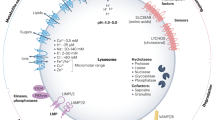
Lysosomal function is performed by lumenal hydrolases that are responsible for substrate digestion and by membrane-associated proteins that handle trafficking of materials into and out of the lysosome.
-
A complex machinery, which includes the kinase complex mammalian target of rapamycin complex 1 (mTORC1, a major regulator of cell growth), the vesicular ATPase complex and additional complexes, is located on the lysosomal surface and is devoted to sensing the nutrient content of the lysosome. This complex is called the lysosomal nutrient sensing (LYNUS) machinery.
-
Most genes encoding lysosomal proteins belong to a gene network termed CLEAR (coordinated lysosomal expression and regulation), and they are transcriptionally regulated by transcription factor EB (TFEB), the master regulator for lysosomal biogenesis. Using this regulatory mechanism, cells can adapt lysosomal function to respond to environmental cues.
-
The activity of TFEB is induced following starvation, by both transcriptional autoregulation and a phosphorylation-dependent mechanism. Once activated, TFEB mediates the starvation response by activating lipid catabolism via the regulation of the master lipid metabolism genes PPARα (peroxisome proliferator-activated receptor-α) and PGC1α (PPARγ co-activator 1α). TFEB regulation and function are conserved in worms.
-
Lysosomal and autophagy dysfunction occurs both in lysosomal storage diseases (LSDs) and in common neurodegenerative diseases, resulting in defective cellular clearance and the accumulation of toxic material. Thus, TFEB-mediated induction of cellular clearance may represent an attractive therapeutic strategy for these disorders.
Abstract
For a long time, lysosomes were considered merely to be cellular 'incinerators' involved in the degradation and recycling of cellular waste. However, now there is compelling evidence indicating that lysosomes have a much broader function and that they are involved in fundamental processes such as secretion, plasma membrane repair, signalling and energy metabolism. Furthermore, the essential role of lysosomes in autophagic pathways puts these organelles at the crossroads of several cellular processes, with significant implications for health and disease. The identification of a master regulator, transcription factor EB (TFEB), that regulates lysosomal biogenesis and autophagy has revealed how the lysosome adapts to environmental cues, such as starvation, and targeting TFEB may provide a novel therapeutic strategy for modulating lysosomal function in human disease.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
de Duve, C. The lysosome turns fifty. Nature Cell Biol. 7, 847–849 (2005).
Saftig, P. & Klumperman, J. Lysosome biogenesis and lysosomal membrane proteins: trafficking meets function. Nature Rev. Mol. Cell Biol. 10, 623–635 (2009). A comprehensive overview of lysosomal function and the role of lysosomal membrane proteins.
Luzio, J. P., Parkinson, M. D., Gray, S. R. & Bright, N. A. The delivery of endocytosed cargo to lysosomes. Biochem. Soc. Trans. 37, 1019–1021 (2009).
Mizushima, N., Levine, B., Cuervo, A. M. & Klionsky, D. J. Autophagy fights disease through cellular self-digestion. Nature 451, 1069–1075 (2008).
Kaushik, S. & Cuervo, A. M. Chaperone-mediated autophagy: a unique way to enter the lysosome world. Trends Cell Biol. 22, 407–417 (2012).
Mijaljica, D., Prescott, M. & Devenish, R. J. Microautophagy in mammalian cells: revisiting a 40-year-old conundrum. Autophagy 7, 673–682 (2011).
Chieregatti, E. & Meldolesi, J. Regulated exocytosis: new organelles for non-secretory purposes. Nature Rev. Mol. Cell Biol. 6, 181–187 (2005).
Verhage, M. & Toonen, R. F. Regulated exocytosis: merging ideas on fusing membranes. Curr. Opin. Cell Biol. 19, 402–408 (2007).
Blott, E. J. & Griffiths, G. M. Secretory lysosomes. Nature Rev. Mol. Cell Biol. 3, 122–131 (2002).
Mostov, K. & Werb, Z. Journey across the osteoclast. Science 276, 219–220 (1997).
Stinchcombe, J., Bossi, G. & Griffiths, G. M. Linking albinism and immunity: the secrets of secretory lysosomes. Science 305, 55–59 (2004).
Sardiello, M. et al. A gene network regulating lysosomal biogenesis and function. Science 325, 473–477 (2009). Discovers that lysosomal function is subject to global transcriptional regulation by the master regulator TFEB.
Settembre, C. et al. TFEB links autophagy to lysosomal biogenesis. Science 332, 1429–1433 (2011). Shows that the biogenesis of both lysosomes and autophagosomes are jointly regulated by TFEB. Starvation induces TFEB cytoplasm-to-nucleus shuttling via a phosphorylation-dependent mechanism.
Braulke, T. & Bonifacino, J. S. Sorting of lysosomal proteins. Biochim. Biophys. Acta 1793, 605–614 (2009).
Luzio, J. P., Pryor, P. R. & Bright, N. A. Lysosomes: fusion and function. Nature Rev. Mol. Cell Biol. 8, 622–632 (2007).
Pfeffer, S. R. Rab GTPases: specifying and deciphering organelle identity and function. Trends Cell Biol. 11, 487–491 (2001).
Zerial, M. & McBride, H. Rab proteins as membrane organizers. Nature Rev. Mol. Cell Biol. 2, 107–117 (2001).
Rink, J., Ghigo, E., Kalaidzidis, Y. & Zerial, M. Rab conversion as a mechanism of progression from early to late endosomes. Cell 122, 735–749 (2005).
Henne, W. M., Buchkovich, N. J. & Emr, S. D. The ESCRT pathway. Dev. Cell 21, 77–91 (2011).
Luzio, J. P. et al. ESCRT proteins and the regulation of endocytic delivery to lysosomes. Biochem. Soc. Trans. 37, 178–180 (2009).
Sridhar, S. et al. The lipid kinase PI4KIIIβ preserves lysosomal identity. EMBO J. 32, 324–339 (2013).
Schulze, H., Kolter, T. & Sandhoff, K. Principles of lysosomal membrane degradation: cellular topology and biochemistry of lysosomal lipid degradation. Biochim. Biophys. Acta 1793, 674–683 (2009).
Rojas, R. et al. Regulation of retromer recruitment to endosomes by sequential action of Rab5 and Rab7. J. Cell Biol. 183, 513–526 (2008).
Wang, T., Ming, Z., Xiaochun, W. & Hong, W. Rab7: role of its protein interaction cascades in endo-lysosomal traffic. Cell. Signal. 23, 516–521 (2011).
Pryor, P. R. et al. Combinatorial SNARE complexes with VAMP7 or VAMP8 define different late endocytic fusion events. EMBO Rep. 5, 590–595 (2004).
Weber, T. et al. SNAREpins: minimal machinery for membrane fusion. Cell 92, 759–772 (1998).
Jahn, R. & Scheller, R. H. SNAREs — engines for membrane fusion. Nature Rev. Mol. Cell Biol. 7, 631–643 (2006).
Ohya, T. et al. Reconstitution of Rab- and SNARE-dependent membrane fusion by synthetic endosomes. Nature 459, 1091–1097 (2009).
Zeigerer, A. et al. Rab5 is necessary for the biogenesis of the endolysosomal system in vivo. Nature 485, 465–470 (2012).
Itakura, E., Kishi-Itakura, C. & Mizushima, N. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell 151, 1256–1269 (2012).
Hamasaki, M. et al. Autophagosomes form at ER–mitochondria contact sites. Nature 495, 389–393 (2013).
Ghosh, P., Dahms, N. M. & Kornfeld, S. Mannose 6-phosphate receptors: new twists in the tale. Nature Rev. Mol. Cell Biol. 4, 202–212 (2003).
Neufeld, E. F. The uptake of enzymes into lysosomes: an overview. Birth Defects Orig. Artic. Ser. 16, 77–84 (1980).
Reczek, D. et al. LIMP-2 is a receptor for lysosomal mannose-6-phosphate-independent targeting of β-glucocerebrosidase. Cell 131, 770–783 (2007). Reveals a new transport mechanism of lysosomal enzymes that is responsible for the targeting of β-glucocerebrosidase.
Gallala, H. D., Breiden, B. & Sandhoff, K. Regulation of the NPC2 protein-mediated cholesterol trafficking by membrane lipids. J. Neurochem. 116, 702–707 (2011).
Munford, R. S., Sheppard, P. O. & O'Hara, P. J. Saposin-like proteins (SAPLIP) carry out diverse functions on a common backbone structure. J. Lipid Res. 36, 1653–1663 (1995).
Kolter, T. & Sandhoff, K. Principles of lysosomal membrane digestion: stimulation of sphingolipid degradation by sphingolipid activator proteins and anionic lysosomal lipids. Annu. Rev. Cell Dev. Biol. 21, 81–103 (2005).
Furst, W. & Sandhoff, K. Activator proteins and topology of lysosomal sphingolipid catabolism. Biochim. Biophys. Acta 1126, 1–16 (1992).
Mobius, W., Herzog, V., Sandhoff, K. & Schwarzmann, G. Intracellular distribution of a biotin-labeled ganglioside, GM1, by immunoelectron microscopy after endocytosis in fibroblasts. J. Histochem. Cytochem. 47, 1005–1014 (1999).
Burkhardt, J. K. et al. Accumulation of sphingolipids in SAP-precursor (prosaposin)-deficient fibroblasts occurs as intralysosomal membrane structures and can be completely reversed by treatment with human SAP-precursor. Eur. J. Cell Biol. 73, 10–18 (1997).
Bradova, V. et al. Prosaposin deficiency: further characterization of the sphingolipid activator protein-deficient sibs. Multiple glycolipid elevations (including lactosylceramidosis), partial enzyme deficiencies and ultrastructure of the skin in this generalized sphingolipid storage disease. Hum. Genet. 92, 143–152 (1993).
Schnabel, D. et al. Simultaneous deficiency of sphingolipid activator proteins 1 and 2 is caused by a mutation in the initiation codon of their common gene. J. Biol. Chem. 267, 3312–3315 (1992).
Cosma, M. P. et al. The multiple sulfatase deficiency gene encodes an essential and limiting factor for the activity of sulfatases. Cell 113, 445–456 (2003).
Dierks, T. et al. Multiple sulfatase deficiency is caused by mutations in the gene encoding the human Cα-formylglycine generating enzyme. Cell 113, 435–444 (2003).
Bagshaw, R. D., Mahuran, D. J. & Callahan, J. W. Lysosomal membrane proteomics and biogenesis of lysosomes. Mol. Neurobiol. 32, 27–41 (2005).
Callahan, J. W., Bagshaw, R. D. & Mahuran, D. J. The integral membrane of lysosomes: its proteins and their roles in disease. J. Proteom. 72, 23–33 (2009).
Lubke, T., Lobel, P. & Sleat, D. E. Proteomics of the lysosome. Biochim. Biophys. Acta 1793, 625–635 (2009).
Schroder, B. A., Wrocklage, C., Hasilik, A. & Saftig, P. The proteome of lysosomes. Proteomics 10, 4053–4076 (2010).
Sleat, D. E., Jadot, M. & Lobel, P. Lysosomal proteomics and disease. Proteomics Clin. Appl. 1, 1134–1146 (2007).
Conner, S. D. & Schmid, S. L. Regulated portals of entry into the cell. Nature 422, 37–44 (2003).
Doherty, G. J. & McMahon, H. T. Mechanisms of endocytosis. Annu. Rev. Biochem. 78, 857–902 (2009).
Hansen, C. G. & Nichols, B. J. Molecular mechanisms of clathrin-independent endocytosis. J. Cell Sci. 122, 1713–1721 (2009).
Sorkin, A. & von Zastrow, M. Endocytosis and signalling: intertwining molecular networks. Nature Rev. Mol. Cell Biol. 10, 609–622 (2009).
Katzmann, D. J., Babst, M. & Emr, S. D. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell 106, 145–155 (2001).
Raiborg, C. & Stenmark, H. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature 458, 445–452 (2009).
Haglund, K. & Dikic, I. The role of ubiquitylation in receptor endocytosis and endosomal sorting. J. Cell Sci. 125, 265–275 (2012).
Ohkuma, S. & Poole, B. Fluorescence probe measurement of the intralysosomal pH in living cells and the perturbation of pH by various agents. Proc. Natl Acad. Sci. USA 75, 3327–3331 (1978).
Ohkuma, S., Moriyama, Y. & Takano, T. Identification and characterization of a proton pump on lysosomes by fluorescein–isothiocyanate–dextran fluorescence. Proc. Natl Acad. Sci. USA 79, 2758–2762 (1982).
Graves, A. R., Curran, P. K., Smith, C. L. & Mindell, J. A. The Cl−/H+ antiporter ClC-7 is the primary chloride permeation pathway in lysosomes. Nature 453, 788–792 (2008).
Kasper, D. et al. Loss of the chloride channel ClC-7 leads to lysosomal storage disease and neurodegeneration. EMBO J. 24, 1079–1091 (2005).
Weinert, S. et al. Lysosomal pathology and osteopetrosis upon loss of H+-driven lysosomal Cl− accumulation. Science 328, 1401–1403 (2010).
Mindell, J. A. Lysosomal acidification mechanisms. Annu. Rev. Physiol. 74, 69–86 (2012).
Zhang, F., Jin, S., Yi, F. & Li, P. L. TRP–ML1 functions as a lysosomal NAADP-sensitive Ca2+ release channel in coronary arterial myocytes. J. Cell. Mol. Med. 13, 3174–3185 (2009).
Zhang, F., Xu, M., Han, W. Q. & Li, P. L. Reconstitution of lysosomal NAADP–TRP–ML1 signaling pathway and its function in TRP–ML1−/− cells. Am. J. Physiol. Cell Physiol. 301, C421–C430 (2011).
Calcraft, P. J. et al. NAADP mobilizes calcium from acidic organelles through two-pore channels. Nature 459, 596–600 (2009).
Sahu, R. et al. Microautophagy of cytosolic proteins by late endosomes. Dev. Cell 20, 131–139 (2011).
Ahlberg, J., Marzella, L. & Glaumann, H. Uptake and degradation of proteins by isolated rat liver lysosomes. Suggestion of a microautophagic pathway of proteolysis. Lab. Invest. 47, 523–532 (1982).
Cuervo, A. M. & Dice, J. F. A receptor for the selective uptake and degradation of proteins by lysosomes. Science 273, 501–503 (1996).
Chiang, H. L., Terlecky, S. R., Plant, C. P. & Dice, J. F. A role for a 70-kilodalton heat shock protein in lysosomal degradation of intracellular proteins. Science 246, 382–385 (1989).
He, C. & Klionsky, D. J. Regulation mechanisms and signaling pathways of autophagy. Annu. Rev. Genet. 43, 67–93 (2009). Reviews the complex molecular mechanisms and pathways involved in the regulation of autophagy.
Ravikumar, B. et al. Regulation of mammalian autophagy in physiology and pathophysiology. Physiol. Rev. 90, 1383–1435 (2010).
Rodriguez, A., Webster, P., Ortego, J. & Andrews, N. W. Lysosomes behave as Ca2+-regulated exocytic vesicles in fibroblasts and epithelial cells. J. Cell Biol. 137, 93–104 (1997).
Chavez, R. A., Miller, S. G. & Moore, H. P. A biosynthetic regulated secretory pathway in constitutive secretory cells. J. Cell Biol. 133, 1177–1191 (1996).
Coorssen, J. R., Schmitt, H. & Almers, W. Ca2+ triggers massive exocytosis in Chinese hamster ovary cells. EMBO J. 15, 3787–3791 (1996).
Stinchcombe, J. C. & Griffiths, G. M. Regulated secretion from hemopoietic cells. J. Cell Biol. 147, 1–6 (1999).
Andrews, N. W. Regulated secretion of conventional lysosomes. Trends Cell Biol. 10, 316–321 (2000).
Jaiswal, J. K., Andrews, N. W. & Simon, S. M. Membrane proximal lysosomes are the major vesicles responsible for calcium-dependent exocytosis in nonsecretory cells. J. Cell Biol. 159, 625–635 (2002).
Stinchcombe, J. C. & Griffiths, G. M. Secretory mechanisms in cell-mediated cytotoxicity. Annu. Rev. Cell Dev. Biol. 23, 495–517 (2007).
Logan, M. R., Odemuyiwa, S. O. & Moqbel, R. Understanding exocytosis in immune and inflammatory cells: the molecular basis of mediator secretion. J. Allergy Clin. Immunol. 111, 923–932 (2003).
Wesolowski, J. & Paumet, F. The impact of bacterial infection on mast cell degranulation. Immunol. Res. 51, 215–226 (2011).
Ren, Q., Ye, S. & Whiteheart, S. W. The platelet release reaction: just when you thought platelet secretion was simple. Curr. Opin. Hematol. 15, 537–541 (2008).
Tulsiani, D. R., Abou-Haila, A., Loeser, C. R. & Pereira, B. M. The biological and functional significance of the sperm acrosome and acrosomal enzymes in mammalian fertilization. Exp. Cell Res. 240, 151–164 (1998).
Rao, S. K., Huynh, C., Proux-Gillardeaux, V., Galli, T. & Andrews, N. W. Identification of SNAREs involved in synaptotagmin VII-regulated lysosomal exocytosis. J. Biol. Chem. 279, 20471–20479 (2004).
Bossi, G. & Griffiths, G. M. CTL secretory lysosomes: biogenesis and secretion of a harmful organelle. Semin. Immunol. 17, 87–94 (2005).
LaPlante, J. M. et al. Lysosomal exocytosis is impaired in mucolipidosis type IV. Mol. Genet. Metab. 89, 339–348 (2006).
Medina, D. L. et al. Transcriptional activation of lysosomal exocytosis promotes cellular clearance. Dev. Cell 21, 421–430 (2011). The first demonstration that TFEB promotes cellular clearance in human disease.
Dong, X. P. et al. Activating mutations of the TRPML1 channel revealed by proline-scanning mutagenesis. J. Biol. Chem. 284, 32040–32052 (2009).
Cadwell, K. et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature 456, 259–263 (2008).
DeSelm, C. J. et al. Autophagy proteins regulate the secretory component of osteoclastic bone resorption. Dev. Cell 21, 966–974 (2011).
Ganesan, A. K. et al. Genome-wide siRNA-based functional genomics of pigmentation identifies novel genes and pathways that impact melanogenesis in human cells. PLoS Genet. 4, e1000298 (2008).
Gerasimenko, J. V., Gerasimenko, O. V. & Petersen, O. H. Membrane repair: Ca2+-elicited lysosomal exocytosis. Curr. Biol. 11, R971–R974 (2001).
Reddy, A., Caler, E. V. & Andrews, N. W. Plasma membrane repair is mediated by Ca2+-regulated exocytosis of lysosomes. Cell 106, 157–169 (2001).
Roy, D. et al. A process for controlling intracellular bacterial infections induced by membrane injury. Science 304, 1515–1518 (2004).
Han, R. et al. Dysferlin-mediated membrane repair protects the heart from stress-induced left ventricular injury. J. Clin. Invest. 117, 1805–1813 (2007).
Ferron, M. et al. A RANKL–PKCβ–TFEB signaling cascade is necessary for lysosomal biogenesis in osteoclasts. Genes Dev. (in the press) (doi:10.1101/gad.213827.113).
Laplante, M. & Sabatini, D. M. mTOR signaling in growth control and disease. Cell 149, 274–293 (2012).
Sancak, Y. et al. Ragulator–Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 141, 290–303 (2010).
Efeyan, A. et al. Regulation of mTORC1 by the Rag GTPases is necessary for neonatal autophagy and survival. Nature 493, 679–183 (2013).
Ganley, I. G. et al. ULK1·ATG13·FIP200 complex mediates mTOR signaling and is essential for autophagy. J. Biol. Chem. 284, 12297–12305 (2009).
Hosokawa, N. et al. Nutrient-dependent mTORC1 association with the ULK1–Atg13–FIP200 complex required for autophagy. Mol. Biol. Cell 20, 1981–1991 (2009).
Jung, C. H. et al. ULK–Atg13–FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol. Biol. Cell 20, 1992–2003 (2009).
Chan, E. Y., Kir, S. & Tooze, S. A. siRNA screening of the kinome identifies ULK1 as a multidomain modulator of autophagy. J. Biol. Chem. 282, 25464–25474 (2007).
Hara, T. et al. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J. Cell Biol. 181, 497–510 (2008).
Zoncu, R. et al. mTORC1 senses lysosomal amino acids through an inside–out mechanism that requires the vacuolar H+-ATPase. Science 334, 678–683 (2011). Identifies an amino acid sensing machinery that is located on the lysosomal surface and involves mTORC1. This implicates the lysosome in signalling and cellular energy metabolism.
Yu, L. et al. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature 465, 942–946 (2010).
Cang, C. et al. mTOR regulates lysosomal ATP-sensitive two-pore Na+ channels to adapt to metabolic state. Cell 152, 778–790 (2013).
Zoncu, R., Efeyan, A. & Sabatini, D. M. mTOR: from growth signal integration to cancer, diabetes and ageing. Nature Rev. Mol. Cell Biol. 12, 21–35 (2011).
Settembre, C. et al. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 31, 1095–1108 (2012).
Rong, Y. et al. Spinster is required for autophagic lysosome reformation and mTOR reactivation following starvation. Proc. Natl Acad. Sci. USA 108, 7826–7831 (2011).
Rong, Y. et al. Clathrin and phosphatidylinositol-4,5-bisphosphate regulate autophagic lysosome reformation. Nature Cell Biol. 14, 924–934 (2012).
Rehli, M., Den Elzen, N., Cassady, A. I., Ostrowski, M. C. & Hume, D. A. Cloning and characterization of the murine genes for bHLH-ZIP transcription factors TFEC and TFEB reveal a common gene organization for all MiT subfamily members. Genomics 56, 111–120 (1999).
Medendorp, K. et al. Molecular mechanisms underlying the MiT translocation subgroup of renal cell carcinomas. Cytogenet. Genome Res. 118, 157–165 (2007).
Ma, X., Godar, R. J., Liu, H. & Diwan, A. Enhancing lysosome biogenesis attenuates BNIP3-induced cardiomyocyte death. Autophagy 8, 297–309 (2012).
Palmieri, M. et al. Characterization of the CLEAR network reveals an integrated control of cellular clearance pathways. Hum. Mol. Genet. 20, 3852–3866 (2011).
Cuervo, A. M. Cell biology. Autophagy's top chef. Science 332, 1392–1393 (2011).
Ma, D., Panda, S. & Lin, J. D. Temporal orchestration of circadian autophagy rhythm by C/EBPβ. EMBO J. 30, 4642–4651 (2011).
Rzymski, T. et al. Regulation of autophagy by ATF4 in response to severe hypoxia. Oncogene 29, 4424–4435 (2010).
Rouschop, K. M. et al. The unfolded protein response protects human tumor cells during hypoxia through regulation of the autophagy genes MAP1LC3B and ATG5. J. Clin. Invest. 120, 127–141 (2010).
van der Vos, K. E. et al. Modulation of glutamine metabolism by the PI(3)K–PKB–FOXO network regulates autophagy. Nature Cell Biol. 14, 829–837 (2012).
Demontis, F. & Perrimon, N. FOXO/4E-BP signaling in Drosophila muscles regulates organism-wide proteostasis during aging. Cell 143, 813–825 (2010).
Zhao, J. et al. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 6, 472–483 (2007).
Mammucari, C. et al. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 6, 458–471 (2007).
Chauhan, S. et al. ZKSCAN3 is a master transcriptional repressor of autophagy. Mol. Cell (2013).
Calnan, D. R. & Brunet, A. The FoxO code. Oncogene 27, 2276–2288 (2008).
Dephoure, N. et al. A quantitative atlas of mitotic phosphorylation. Proc. Natl Acad. Sci. USA 105, 10762–10767 (2008).
Cea, M. et al. Targeting NAD+ salvage pathway induces autophagy in multiple myeloma cells via mTORC1 and extracellular signal-regulated kinase (ERK1/2) inhibition. Blood 120, 3519–3529 (2012).
Martina, J. A., Chen, Y., Gucek, M. & Puertollano, R. mTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy 8, 903–914 (2012).
Pena-Llopis, S. et al. Regulation of TFEB and V-ATPases by mTORC1. EMBO J. 30, 3242–3258 (2011).
Roczniak-Ferguson, A. et al. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci. Signal. 5, ra42 (2012).
Martina, J. A. & Puertollano, R. Rag GTPases mediate amino acid-dependent recruitment of TFEB and MITF to lysosomes. J. Cell Biol. 200, 475–491 (2013).
Settembre, C. et al. TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nature Cell Biol. 21 Apr 2013 (doi:10.1038/ncb2718).
Singh, R. & Cuervo, A. M. Autophagy in the cellular energetic balance. Cell Metab. 13, 495–504 (2011). Shows that lipid droplets are sequestered by autophagosomes for degradation and recycling to generate free fatty acids.
Singh, R. et al. Autophagy regulates lipid metabolism. Nature 458, 1131–1135 (2009).
Rodriguez-Navarro, J. A. & Cuervo, A. M. Dietary lipids and aging compromise chaperone-mediated autophagy by similar mechanisms. Autophagy 8, 1152–1154 (2012).
Rodriguez-Navarro, J. A. et al. Inhibitory effect of dietary lipids on chaperone-mediated autophagy. Proc. Natl Acad. Sci. USA 109, e705–e714 (2012).
Yang, L., Li, P., Fu, S., Calay, E. S. & Hotamisligil, G. S. Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell Metab. 11, 467–478 (2010).
Woloszynek, J. C., Coleman, T., Semenkovich, C. F. & Sands, M. S. Lysosomal dysfunction results in altered energy balance. J. Biol. Chem. 282, 35765–35771 (2007).
Du, H., Duanmu, M., Witte, D. & Grabowski, G. A. Targeted disruption of the mouse lysosomal acid lipase gene: long-term survival with massive cholesteryl ester and triglyceride storage. Hum. Mol. Genet. 7, 1347–1354 (1998).
Finck, B. N. & Kelly, D. P. PGC-1 coactivators: inducible regulators of energy metabolism in health and disease. J. Clin. Invest. 116, 615–622 (2006).
Grove, C. A. et al. A multiparameter network reveals extensive divergence between C. elegans bHLH transcription factors. Cell 138, 314–327 (2009).
O'Rourke, E. J. & Ruvkun, G. MXL-3 and HLH-30 transcriptionally link lysosomal lipolysis and autophagy to nutrient availability. Nature Cell Biol. 21 Apr 2013 (doi:10.1038/ncb2741).
Kaeberlein, T. L. et al. Lifespan extension in Caenorhabditis elegans by complete removal of food. Aging Cell 5, 487–494 (2006).
Melendez, A. et al. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science 301, 1387–1391 (2003).
Steingrimsson, E., Tessarollo, L., Reid, S. W., Jenkins, N. A. & Copeland, N. G. The bHLH-Zip transcription factor Tfeb is essential for placental vascularization. Development 125, 4607–4616 (1998).
Cuervo, A. M. & Dice, J. F. When lysosomes get old. Exp. Gerontol. 35, 119–131 (2000).
Rubinsztein, D. C., Marino, G. & Kroemer, G. Autophagy and aging. Cell 146, 682–695 (2011).
Ballabio, A. & Gieselmann, V. Lysosomal disorders: from storage to cellular damage. Biochim. Biophys. Acta 1793, 684–696 (2009).
Cox, T. M. & Cachon-Gonzalez, M. B. The cellular pathology of lysosomal diseases. J. Pathol. 226, 241–254 (2012).
Futerman, A. H. & van Meer, G. The cell biology of lysosomal storage disorders. Nature Rev. Mol. Cell Biol. 5, 554–565 (2004).
Schultz, M. L., Tecedor, L., Chang, M. & Davidson, B. L. Clarifying lysosomal storage diseases. Trends Neurosciences 34, 401–410 (2011).
Vitner, E. B., Platt, F. M. & Futerman, A. H. Common and uncommon pathogenic cascades in lysosomal storage diseases. J. Biol. Chem. 285, 20423–20427 (2010).
Walkley, S. U. Pathogenic cascades in lysosomal disease — why so complex? J. Inherit. Metab. Dis. 32, 181–189 (2009).
Lieberman, A. P. et al. Autophagy in lysosomal storage disorders. Autophagy 8, 719–730 (2012).
de Pablo-Latorre, R. et al. Impaired parkin-mediated mitochondrial targeting to autophagosomes differentially contributes to tissue pathology in lysosomal storage diseases. Hum. Mol. Genet. 21, 1770–1781 (2012).
Di Malta, C., Fryer, J. D., Settembre, C. & Ballabio, A. Astrocyte dysfunction triggers neurodegeneration in a lysosomal storage disorder. Proc. Natl Acad. Sci. USA 109, E2334–E2342 (2012).
Fraldi, A. et al. Lysosomal fusion and SNARE function are impaired by cholesterol accumulation in lysosomal storage disorders. EMBO J. 29, 3607–3620 (2010).
Settembre, C. et al. A block of autophagy in lysosomal storage disorders. Hum. Mol. Genet. 17, 119–129 (2008).
Cox, T. M. in Lysosomal Storage Disorders: A Practical Guide (ed. Atul Mehta, B. W. ) 153–165 (Wiley-Blackwell, 2012).
Wong, E. & Cuervo, A. M. Autophagy gone awry in neurodegenerative diseases. Nature Neurosci. 13, 805–811 (2010).
Harris, H. & Rubinsztein, D. C. Control of autophagy as a therapy for neurodegenerative disease. Nature Rev. Neurol. 8, 108–117 (2012). A comprehensive overview of how the modulation of autophagy can be a promising therapeutic strategy for neurodegenerative diseases.
Jeong, H. et al. Acetylation targets mutant huntingtin to autophagosomes for degradation. Cell 137, 60–72 (2009).
Cuervo, A. M., Stefanis, L., Fredenburg, R., Lansbury, P. T. & Sulzer, D. Impaired degradation of mutant α-synuclein by chaperone-mediated autophagy. Science 305, 1292–1295 (2004).
Winslow, A. R. et al. α-synuclein impairs macroautophagy: implications for Parkinson's disease. J. Cell Biol. 190, 1023–1037 (2010).
Orenstein, S. J. et al. Interplay of LRRK2 with chaperone-mediated autophagy. Nature Neurosci. 16, 394–406 (2013).
Martinez-Vicente, M. et al. Cargo recognition failure is responsible for inefficient autophagy in Huntington's disease. Nature Neurosci. 13, 567–576 (2010).
Aharon-Peretz, J., Rosenbaum, H. & Gershoni-Baruch, R. Mutations in the glucocerebrosidase gene and Parkinson's disease in Ashkenazi Jews. N. Engl. J. Med. 351, 1972–1977 (2004).
Sidransky, E. et al. Multicenter analysis of glucocerebrosidase mutations in Parkinson's disease. N. Engl. J. Med. 361, 1651–1661 (2009).
Brady, R. O., Kanfer, J. N. & Shapiro, D. Metabolism of glucocerebrosides. II. Evidence of an enzymatic deficiency in Gaucher's disease. Biochem. Biophys. Res. Commun. 18, 221–225 (1965).
Mazzulli, J. R. et al. Gaucher disease glucocerebrosidase and α-synuclein form a bidirectional pathogenic loop in synucleinopathies. Cell 146, 37–52 (2011). Demonstrates how partial deficiency of the lysosomal enzyme β-glucocerebrosidase can be a major predisposing factor in Parkinson's disease.
Ramirez, A. et al. Hereditary parkinsonism with dementia is caused by mutations in ATP13A2, encoding a lysosomal type 5 P-type ATPase. Nature Genet. 38, 1184–1191 (2006).
Usenovic, M. & Krainc, D. Lysosomal dysfunction in neurodegeneration: the role of ATP13A2/PARK9. Autophagy 8, 987–988 (2012).
Valente, E. M. et al. Hereditary early-onset Parkinson's disease caused by mutations in PINK1. Science 304, 1158–1160 (2004).
Geisler, S. et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nature Cell Biol. 12, 119–131 (2010).
Narendra, D., Tanaka, A., Suen, D. F. & Youle, R. J. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 183, 795–803 (2008).
Kitada, T. et al. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature 392, 605–608 (1998).
Zimprich, A. et al. A mutation in VPS35, encoding a subunit of the retromer complex, causes late-onset Parkinson disease. Am. J. Hum. Genet. 89, 168–175 (2011).
Chartier-Harlin, M. C. et al. Translation initiator EIF4G1 mutations in familial Parkinson disease. Am. J. Hum. Genet. 89, 398–406 (2011).
Lee, J. H. et al. Lysosomal proteolysis and autophagy require presenilin 1 and are disrupted by Alzheimer-related PS1 mutations. Cell 141, 1146–1158 (2010). An important example of lysosomal dysfunction associated with Alzheimer's disease.
Coen, K. et al. Lysosomal calcium homeostasis defects, not proton pump defects, cause endo–lysosomal dysfunction in PSEN-deficient cells. J. Cell Biol. 198, 23–35 (2012).
Skibinski, G. et al. Mutations in the endosomal ESCRTIII-complex subunit CHMP2B in frontotemporal dementia. Nature Genet. 37, 806–808 (2005).
Verhoeven, K. et al. Mutations in the small GTP-ase late endosomal protein RAB7 cause Charcot–Marie–Tooth type 2B neuropathy. Am. J. Hum. Genet. 72, 722–727 (2003).
Saitsu, H. et al. De novo mutations in the autophagy gene WDR45 cause static encephalopathy of childhood with neurodegeneration in adulthood. Nature Genet. 45, 445–449 (2013).
Yang, D. S. et al. Reversal of autophagy dysfunction in the TgCRND8 mouse model of Alzheimer's disease ameliorates amyloid pathologies and memory deficits. Brain 134, 258–277 (2011).
Mueller-Steiner, S. et al. Antiamyloidogenic and neuroprotective functions of cathepsin B: implications for Alzheimer's disease. Neuron 51, 703–714 (2006).
Sun, B. et al. Cystatin C–cathepsin B axis regulates amyloid-β levels and associated neuronal deficits in an animal model of Alzheimer's disease. Neuron 60, 247–257 (2008).
Ravikumar, B. et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nature Genet. 36, 585–595 (2004).
Menzies, F. M. et al. Autophagy induction reduces mutant ataxin-3 levels and toxicity in a mouse model of spinocerebellar ataxia type 3. Brain 133, 93–104 (2010).
Rose, C. et al. Rilmenidine attenuates toxicity of polyglutamine expansions in a mouse model of Huntington's disease. Hum. Mol. Genet. 19, 2144–2153 (2010).
Tanaka, M. et al. Trehalose alleviates polyglutamine-mediated pathology in a mouse model of Huntington disease. Nature Med. 10, 148–154 (2004).
Spampanato, C. et al. Transcription factor EB (TFEB) is a new therapeutic target for Pompe disease. EMBO Mol Med. 11 Apr 2013 (doi:10.1002/emmm.201202176.
Dehay, B. et al. Pathogenic lysosomal depletion in Parkinson's disease. J. Neurosci. 30, 12535–12544 (2010).
Tsunemi, T. et al. PGC-1α rescues Huntington's disease proteotoxicity by preventing oxidative stress and promoting TFEB function. Sci. Transl. Med. 4, 142ra97 (2012).
Pastore, N. et al. Gene transfer of master autophagy regulator TFEB results in clearance of toxic protein and correction of hepatic disease in α-1-anti-trypsin deficiency. EMBO Mol. Med. 5, 397–412 (2013).
Bagshaw, R. D., Mahuran, D. J. & Callahan, J. W. A proteomic analysis of lysosomal integral membrane proteins reveals the diverse composition of the organelle. Mol. Cell. Proteom. 4, 133–143 (2005).
Kobayashi, T. et al. Separation and characterization of late endosomal membrane domains. J. Biol. Chem. 277, 32157–32164 (2002).
Andrejewski, N. et al. Normal lysosomal morphology and function in LAMP-1-deficient mice. J. Biol. Chem. 274, 12692–12701 (1999).
Nishi, T. & Forgac, M. The vacuolar (H+)-ATPases — nature's most versatile proton pumps. Nature Rev. Mol. Cell Biol. 3, 94–103 (2002).
Marshansky, V. & Futai, M. The V-type H+-ATPase in vesicular trafficking: targeting, regulation and function. Curr. Opin. Cell Biol. 20, 415–426 (2008).
Dong, X. P. et al. The type IV mucolipidosis-associated protein TRPML1 is an endolysosomal iron release channel. Nature 455, 992–996 (2008).
Wong, C. O., Li, R., Montell, C. & Venkatachalam, K. Drosophila TRPML is required for TORC1 activation. Curr. Biol. 22, 1616–1621 (2012).
Bargal, R. et al. Identification of the gene causing mucolipidosis type IV. Nature Genet. 26, 118–123 (2000).
Bassi, M. T. et al. Cloning of the gene encoding a novel integral membrane protein, mucolipidin-and identification of the two major founder mutations causing mucolipidosis type IV. Am. J. Hum. Genet. 67, 1110–1120 (2000).
Jentsch, T. J., Poet, M., Fuhrmann, J. C. & Zdebik, A. A. Physiological functions of CLC Cl− channels gleaned from human genetic disease and mouse models. Annu. Rev. Physiol. 67, 779–807 (2005).
Cuervo, A. M., Gomes, A. V., Barnes, J. A. & Dice, J. F. Selective degradation of annexins by chaperone-mediated autophagy. J. Biol. Chem. 275, 33329–33335 (2000).
Nishino, I. et al. Primary LAMP-2 deficiency causes X-linked vacuolar cardiomyopathy and myopathy (Danon disease). Nature 406, 906–910 (2000).
Lloyd-Evans, E. et al. Niemann–Pick disease type C1 is a sphingosine storage disease that causes deregulation of lysosomal calcium. Nature Med. 14, 1247–1255 (2008).
Liu, B., Du, H., Rutkowski, R., Gartner, A. & Wang, X. LAAT-1 is the lysosomal lysine/arginine transporter that maintains amino acid homeostasis. Science 337, 351–354 (2012).
Hrebicek, M. et al. Mutations in TMEM76* cause mucopolysaccharidosis IIIC (Sanfilippo C syndrome). Am. J. Hum. Genet. 79, 807–819 (2006).
Fan, X. et al. Identification of the gene encoding the enzyme deficient in mucopolysaccharidosis IIIC (Sanfilippo disease type C). Am. J. Hum. Genet. 79, 738–744 (2006).
Durand, S., Feldhammer, M., Bonneil, E., Thibault, P. & Pshezhetsky, A. V. Analysis of the biogenesis of heparan sulfate acetyl-CoA:α-glucosaminide N-acetyltransferase provides insights into the mechanism underlying its complete deficiency in mucopolysaccharidosis IIIC. J. Biol. Chem. 285, 31233–31242 (2010).
Efeyan, A., Zoncu, R. & Sabatini, D. M. Amino acids and mTORC1: from lysosomes to disease. Trends Mol. Med. 18, 524–533 (2012).
Sancak, Y. et al. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 320, 1496–1501 (2008).
Bar-Peled, L., Schweitzer, L. D., Zoncu, R. & Sabatini, D. M. Ragulator is a GEF for the Rag GTPases that signal amino acid levels to mTORC1. Cell 150, 1196–1208 (2012).
Inoki, K., Li, Y., Xu, T. & Guan, K. L. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 17, 1829–1834 (2003).
Tee, A. R., Manning, B. D., Roux, P. P., Cantley, L. C. & Blenis, J. Tuberous sclerosis complex gene products, Tuberin and Hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Curr. Biol. 13, 1259–1268 (2003).
Acknowledgements
The authors thank T. Braulke, A. De Matteis, J. Irazoqui, D. Rubinsztein, D. Sabatini, P. Saftig and R. Zoncu for the helpful suggestions, G. Diez-Roux for helpful discussions and support during manuscript preparation and E. Abrams for manuscript editing. They acknowledge the support of the Italian Telethon Foundation (TGM11CB6, to C.S. and A.B), the Beyond Batten Disease Foundation (to C.S. and A.B.), the European Research Council Advanced Investigator (250154, to A.B.), March of Dimes (#6-FY11-306, to A.B) and the US National Institutes of Health (R01-NS078072, to A.B).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Glossary
- Glycocalyx
-
The polysaccharide-based coating on the inner side of a lysosomal membrane that protects this organelle from digestion by lysosomal enzymes.
- Autophagosomes
-
Intracytoplasmic vacuoles that contain elements of the cytoplasm of a cell. They fuse with lysosomes, and the contents are subjected to enzymatic digestion.
- Danon disease
-
An X-linked dominant disorder caused by mutations in the gene encoding lysosome-associated membrane protein 2 (LAMP2). It predominantly affects cardiac muscle.
- Niemann–Pick disease type C1
-
An autosomal recessive lipid storage disorder that is caused by mutation in the NPC1 (Niemann Pick type C1) gene. It is characterized by progressive neurodegeneration.
- Lysosome-related organelles
-
(LROs). Cell type-specific compartments that include melanosomes, lytic granules, major histocompatibility complex class II compartments, platelet-dense granules, basophil granules, azurophil granules and Drosophila melanogaster pigment granules.
- Wolman's disease
-
An early-onset fulminant disorder of infancy with substantial infiltration of several organs, including the spleen and the liver, by macrophages filled with cholesteryl esters and triglycerides. It is caused by mutations in the gene encoding lipase A.
- Multiple sulphatase deficiency
-
(MSD). An autosomal recessive inherited disease that is caused by mutations in the sulphatase-modifying factor 1 (SUMF1) gene.
- Mucopolysaccharidosis
-
(MPS). A metabolic disorder that is caused by the absence or malfunctioning of lysosomal enzymes needed to break down molecules.
- Gaucher's disease
-
An autosomal recessive lysosomal storage disorder due to the deficient activity of β-glucocerebrosidase.
- Fronto-temporal dementia
-
A disorder associated with fronto-temporal lobar degeneration.
- Charcot–Marie–Tooth type 2B
-
Autosomal dominant peripheral sensory neuropathy due to mutations in the late endosomal small GTPase RAB7.
- Neuronal ceroid lipofuscinosis
-
A clinically and genetically heterogeneous group of neurodegenerative disorders that are characterized by the intracellular accumulation of autofluorescent lipopigment storage material.
- Pompe's disease
-
An autosomal recessive inherited disease, also known as glycogen storage disease II. This prototypic lysosomal storage disease is caused by mutations in the gene encoding acid α-1,4-glucosidase.
Rights and permissions
About this article
Cite this article
Settembre, C., Fraldi, A., Medina, D. et al. Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nat Rev Mol Cell Biol 14, 283–296 (2013). https://doi.org/10.1038/nrm3565
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrm3565
This article is cited by
-
TFEB–vacuolar ATPase signaling regulates lysosomal function and microglial activation in tauopathy
Nature Neuroscience (2024)
-
The cytotoxicity of microcystin-LR: ultrastructural and functional damage of cells
Archives of Toxicology (2024)
-
Transcriptome analysis of the immunoprotective effect of Bacillus licheniformis on the intestinal tract of Carassius auratus gibelio infected with Aeromonas hydrophila
Aquaculture International (2024)
-
Superwettable interface towards biodetection in confined space
Nano Research (2024)
-
Reciprocal effects of alpha-synuclein aggregation and lysosomal homeostasis in synucleinopathy models
Translational Neurodegeneration (2023)