Key Points
-
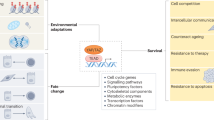
The forkhead box O (FOXO) family of transcription factors consists of four members: FOXO1, FOXO3, FOXO4 and FOXO6. At the cellular level, FOXO transcriptional targets are involved in the regulation of cell cycle, apoptosis, oxidative stress resistance and metabolism.
-
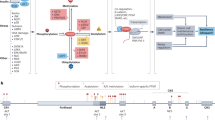
FOXOs integrate signals from multiple upstream pathways and are activated by metabolic and oxidative stress and by the absence of growth factors. This regulation of FOXO activity is accomplished through various post-translational modifications, nuclear–cytoplasmic localization of FOXOs and their regulators, specific microRNAs (miRNAs) and interactions with other transcription factors.
-
The biological role of FOXOs is predominately to respond to stress conditions rather then being an essential mediator of normal physiology. Generally, by responding to and counteracting environmental changes, FOXOs act to maintain homeostasis.
-
In model organisms, FOXO function is diverse and mostly comes into focus under conditions that disturb homeostasis. Conditional knockout models have revealed functions in tumour suppression, metabolic stress and protein homeostasis, stem cell maintenance as well as specific roles in the immune system.
-
The role of FOXOs in maintaining cellular homeostasis reveals many similarities with the role of p53 in maintaining genome homeostasis, and these factors share co-regulators.
-
FOXOs were initially characterized as tumour suppressors. However, FOXOs have also been shown to act as tumour promoters. This seemingly paradoxical outcome of being both tumour suppressor and tumour promoter can be reconciled by postulating that FOXOs act as homeostasis regulators of both normal and cancer cells.
Abstract
Forkhead box O (FOXO) transcription factors are involved in the regulation of the cell cycle, apoptosis and metabolism. In model organisms, FOXO activity also affects stem cell maintenance and lifespan as well as age-related diseases, such as cancer and diabetes. Multiple upstream pathways regulate FOXO activity through post-translational modifications and nuclear–cytoplasmic shuttling of both FOXO and its regulators. The diversity of this upstream regulation and the downstream effects of FOXOs suggest that they function as homeostasis regulators to maintain tissue homeostasis over time and coordinate a response to environmental changes, including growth factor deprivation, metabolic stress (starvation) and oxidative stress.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
van der Horst, A. & Burgering, B. M. Stressing the role of FoxO proteins in lifespan and disease. Nature Rev. Mol. Cell Biol. 8, 440–450 (2007).
Dansen, T. B. & Burgering, B. M. Unravelling the tumor-suppressive functions of FOXO proteins. Trends Cell Biol. 18, 421–429 (2008).
Kenyon, C. J. The genetics of ageing. Nature 464, 504–512 (2010).
Paik, J. H. et al. FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell 128, 309–323 (2007). The first description of mice with conditional deletion of Foxo1, Foxo3 and Foxo4 . Shows that FOXOs can act as tumour suppressors.
Bridge, D. et al. FoxO and stress responses in the cnidarian Hydra vulgaris. PLoS ONE 5, e11686 (2010).
van den Berg, M. C. & Burgering, B. M. Integrating opposing signals toward forkhead box O. Antioxid. Redox Signal. 14, 607–621 (2011).
Calnan, D. R. & Brunet, A. The FoxO code. Oncogene 27, 2276–2288 (2008).
Hardie, D. G., Ross, F. A. & Hawley, S. A. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nature Rev. Mol. Cell Biol. 13, 251–262 (2012).
Greer, E. L. et al. The energy sensor AMP-activated protein kinase directly regulates the mammalian FOXO3 transcription factor. J. Biol. Chem. 282, 30107–30119 (2007).
Greer, E. L. et al. An AMPK–FOXO pathway mediates longevity induced by a novel method of dietary restriction in C. elegans. Curr. Biol. 17, 1646–1656 (2007).
Wang, F. et al. Structures of KIX domain of CBP in complex with two FOXO3a transactivation domains reveal promiscuity and plasticity in coactivator recruitment. Proc. Natl Acad. Sci. USA 109, 6078–6083 (2012).
Huang, H., Regan, K. M., Lou, Z., Chen, J. & Tindall, D. J. CDK2-dependent phosphorylation of FOXO1 as an apoptotic response to DNA damage. Science 314, 294–297 (2006).
Yuan, Z. et al. Activation of FOXO1 by Cdk1 in cycling cells and postmitotic neurons. Science 319, 1665–1668 (2008).
Liu, P., Kao, T. P. & Huang, H. CDK1 promotes cell proliferation and survival via phosphorylation and inhibition of FOXO1 transcription factor. Oncogene 27, 4733–4744 (2008).
Yata, K. & Esashi, F. Dual role of CDKs in DNA repair: to be, or not to be. DNA Repair (Amst.) 8, 6–18 (2009).
Matsuoka, S. et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 316, 1160–1166 (2007).
Tsai, W. B., Chung, Y. M., Takahashi, Y., Xu, Z. & Hu, M. C. Functional interaction between FOXO3a and ATM regulates DNA damage response. Nature Cell Biol. 10, 460–467 (2008).
Yalcin, S. et al. Foxo3 is essential for the regulation of ataxia telangiectasia mutated and oxidative stress-mediated homeostasis of hematopoietic stem cells. J. Biol. Chem. 283, 25692–25705 (2008).
Kress, T. R. et al. The MK5/PRAK kinase and Myc form a negative feedback loop that is disrupted during colorectal tumorigenesis. Mol. Cell 41, 445–457 (2011).
Nakae, J. et al. Novel repressor regulates insulin sensitivity through interaction with Foxo1. EMBO J. 31, 2275–2295 (2012).
Yang, Y., Hou, H., Haller, E. M., Nicosia, S. V. & Bai, W. Suppression of FOXO1 activity by FHL2 through SIRT1-mediated deacetylation. EMBO J. 24, 1021–1032 (2005).
Wang, Y. C. & Li, C. Evolutionarily conserved protein arginine methyltransferases in non-mammalian animal systems. FEBS J. 279, 932–945 (2012).
Yamagata, K. et al. Arginine methylation of FOXO transcription factors inhibits their phosphorylation by Akt. Mol. Cell 32, 221–231 (2008).
Takahashi, Y. et al. Asymmetric arginine dimethylation determines life span in C. elegans by regulating forkhead transcription factor DAF-16. Cell Metab. 13, 505–516 (2011).
Xie, Q. et al. Lysine methylation of FOXO3 regulates oxidative stress-induced neuronal cell death. EMBO Rep. 13, 371–377 (2012).
Housley, M. P. et al. O-GlcNAc regulates FoxO activation in response to glucose. J. Biol. Chem. 283, 16283–16292 (2008).
Kuo, M., Zilberfarb, V., Gangneux, N., Christeff, N. & Issad, T. O-GlcNAc modification of FoxO1 increases its transcriptional activity: a role in the glucotoxicity phenomenon? Biochimie 90, 679–685 (2008).
Puigserver, P. et al. Insulin-regulated hepatic gluconeogenesis through FOXO1–PGC-1α interaction. Nature 423, 550–555 (2003).
Housley, M. P. et al. A PGC-1α–O-GlcNAc transferase complex regulates FoxO transcription factor activity in response to glucose. J. Biol. Chem. 284, 5148–5157 (2009).
Hanover, J. A., Krause, M. W. & Love, D. C. Bittersweet memories: linking metabolism to epigenetics through O-GlcNAcylation. Nature Rev. Mol. Cell Biol. 13, 312–321 (2012).
Dansen, T. B. et al. Redox-sensitive cysteines bridge p300/CBP-mediated acetylation and FoxO4 activity. Nature Chem. Biol. 5, 664–672 (2009). Demonstrates that FOXOs can act as direct sensors of cellular redox.
Szypowska, A. A. & Burgering, B. M. The peroxide dilemma: opposing and mediating insulin action. Antioxid. Redox Signal. 15, 219–232 (2011).
Dyson, H. J. & Wright, P. E. Intrinsically unstructured proteins and their functions. Nature Rev. Mol. Cell Biol. 6, 197–208 (2005).
Lacy, E. R. et al. p27 binds cyclin–CDK complexes through a sequential mechanism involving binding-induced protein folding. Nature Struct. Mol. Biol. 11, 358–364 (2004).
Wang, Y. et al. Intrinsic disorder mediates the diverse regulatory functions of the Cdk inhibitor p21. Nature Chem. Biol. 7, 214–221 (2011).
Mittag, T. et al. Dynamic equilibrium engagement of a polyvalent ligand with a single-site receptor. Proc. Natl Acad. Sci. USA 105, 17772–17777 (2008).
Rena, G., Bain, J., Elliott, M. & Cohen, P. D4476, a cell-permeant inhibitor of CK1, suppresses the site-specific phosphorylation and nuclear exclusion of FOXO1a. EMBO Rep. 5, 60–65 (2004).
Woods, Y. L. et al. The kinase DYRK1A phosphorylates the transcription factor FKHR at Ser329 in vitro, a novel in vivo phosphorylation site. Biochem. J. 355, 597–607 (2001).
Giannakou, M. E. & Partridge, L. The interaction between FOXO and SIRT1: tipping the balance towards survival. Trends Cell Biol. 14, 408–412 (2004).
Brunet, A. et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science 303, 2011–2015 (2004).
Banks, A. S. et al. Dissociation of the glucose and lipid regulatory functions of FoxO1 by targeted knockin of acetylation-defective alleles in mice. Cell Metab. 14, 587–597 (2011).
van der Horst, A. et al. FOXO4 transcriptional activity is regulated by monoubiquitination and USP7/HAUSP. Nature Cell Biol. 8, 1064–1073 (2006).
Oshikawa, K., Matsumoto, M., Oyamada, K. & Nakayama, K. I. Proteome-wide identification of ubiquitylation sites by conjugation of engineered lysine-less ubiquitin. J. Proteome Res. 11, 796–807 (2012).
Asada, S. et al. Mitogen-activated protein kinases, Erk and p38, phosphorylate and regulate Foxo1. Cell. Signal. 19, 519–527 (2007).
Guttilla, I. K. & White, B. A. Coordinate regulation of FOXO1 by miR-27a, miR-96, and miR-182 in breast cancer cells. J. Biol. Chem. 284, 23204–23216 (2009).
Myatt, S. S. et al. Definition of microRNAs that repress expression of the tumor suppressor gene FOXO1 in endometrial cancer. Cancer Res. 70, 367–377 (2010).
Hasseine, L. K. et al. miR-139 impacts FoxO1 action by decreasing FoxO1 protein in mouse hepatocytes. Biochem. Biophys. Res. Commun. 390, 1278–1282 (2009).
Small, E. M. et al. Regulation of PI3-kinase/Akt signaling by muscle-enriched microRNA-486. Proc. Natl Acad. Sci. USA 107, 4218–4223 (2010).
Yamamoto, M. et al. miR-155, a modulator of FOXO3a protein expression, is underexpressed and cannot be upregulated by stimulation of HOZOT, a line of multifunctional treg. PLoS ONE 6, e16841 (2011).
Kong, W. et al. MicroRNA-155 regulates cell survival, growth, and chemosensitivity by targeting FOXO3a in breast cancer. J. Biol. Chem. 285, 17869–17879 (2010).
Liu, X. et al. microRNA-499-5p promotes cellular invasion and tumor metastasis in colorectal cancer by targeting FOXO4 and PDCD4. Carcinogenesis 32, 1798–1805 (2011).
Wang, K. & Li, P. F. Foxo3a regulates apoptosis by negatively targeting miR-21. J. Biol. Chem. 285, 16958–16966 (2010).
Gan, B. et al. FoxOs enforce a progression checkpoint to constrain mTORC1-activated renal tumorigenesis. Cancer Cell 18, 472–484 (2010). Demonstrates, together with reference 99, the relevance of MYC–FOXO antagonism in tumorigenesis.
Brett, J. O., Renault, V. M., Rafalski, V. A., Webb, A. E. & Brunet, A. The microRNA cluster miR-106b∼25 regulates adult neural stem/progenitor cell proliferation and neuronal differentiation. Aging (Albany NY) 3, 108–124 (2011).
Perl, A. Emerging new pathways of pathogenesis and targets for treatment in systemic lupus erythematosus and Sjogren's syndrome. Curr. Opin. Rheumatol. 21, 443–447 (2009).
Dai, R. et al. Identification of a common lupus disease-associated microRNA expression pattern in three different murine models of lupus. PLoS ONE 5, e14302 (2010).
Tanno, M., Sakamoto, J., Miura, T., Shimamoto, K. & Horio, Y. Nucleocytoplasmic shuttling of the NAD+-dependent histone deacetylase SIRT1. J. Biol. Chem. 282, 6823–6832 (2007).
Mihaylova, M. M. et al. Class IIa histone deacetylases are hormone-activated regulators of FOXO and mammalian glucose homeostasis. Cell 145, 607–621 (2011).
Wang, B. et al. A hormone-dependent module regulating energy balance. Cell 145, 596–606 (2011).
Kitamura, Y. I. et al. FoxO1 protects against pancreatic β-cell failure through NeuroD and MafA induction. Cell Metab. 2, 153–163 (2005).
Zhong, S., Salomoni, P. & Pandolfi, P. P. The transcriptional role of PML and the nuclear body. Nature Cell Biol. 2, e85–e90 (2000).
Brenkman, A. B., de Keizer, P. L., van den Broek, N. J., Jochemsen, A. G. & Burgering, B. M. Mdm2 induces mono-ubiquitination of FOXO4. PLoS ONE 3, e2819 (2008).
Yang, J. Y. et al. ERK promotes tumorigenesis by inhibiting FOXO3a via MDM2-mediated degradation. Nature Cell Biol. 10, 138–148 (2008).
Fu, W. et al. MDM2 acts downstream of p53 as an E3 ligase to promote FOXO ubiquitination and degradation. J. Biol. Chem. 284, 13987–14000 (2009).
Ashcroft, M. et al. Phosphorylation of HDM2 by Akt. Oncogene 21, 1955–1962 (2002).
Mayo, L. D. & Donner, D. B. A phosphatidylinositol 3-kinase/Akt pathway promotes translocation of Mdm2 from the cytoplasm to the nucleus. Proc. Natl Acad. Sci. USA 98, 11598–11603 (2001).
Zhou, B. P. et al. HER-2/neu induces p53 ubiquitination via Akt-mediated MDM2 phosphorylation. Nature Cell Biol. 3, 973–982 (2001).
Tao, W. & Levine, A. J. P19ARF stabilizes p53 by blocking nucleo–cytoplasmic shuttling of Mdm2. Proc. Natl Acad. Sci. USA 96, 6937–6941 (1999).
Bernardi, R. et al. PML regulates p53 stability by sequestering Mdm2 to the nucleolus. Nature Cell Biol. 6, 665–672 (2004).
Kurki, S., Latonen, L. & Laiho, M. Cellular stress and DNA damage invoke temporally distinct Mdm2, p53 and PML complexes and damage-specific nuclear relocalization. J. Cell Sci. 116, 3917–3925 (2003).
Jackson, M. W. et al. Hdm2 nuclear export, regulated by insulin-like growth factor-I/MAPK/p90Rsk signaling, mediates the transformation of human cells. J. Biol. Chem. 281, 16814–16820 (2006).
Wagner, S. A. et al. A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol. Cell. Proteomics 10, M111.013284 (2011).
Aoki, M., Jiang, H. & Vogt, P. K. Proteasomal degradation of the FoxO1 transcriptional regulator in cells transformed by the P3k and Akt oncoproteins. Proc. Natl Acad. Sci. USA 101, 13613–13617 (2004).
Meier, R., Alessi, D. R., Cron, P., Andjelkovic, M. & Hemmings, B. A. Mitogenic activation, phosphorylation, and nuclear translocation of protein kinase Bβ. J. Biol. Chem. 272, 30491–30497 (1997).
Brownawell, A. M., Kops, G. J., Macara, I. G. & Burgering, B. M. Inhibition of nuclear import by protein kinase B (Akt) regulates the subcellular distribution and activity of the forkhead transcription factor AFX. Mol. Cell. Biol. 21, 3534–3546 (2001).
Brunet, A. et al. 14-3-3 transits to the nucleus and participates in dynamic nucleocytoplasmic transport. J. Cell Biol. 156, 817–828 (2002).
Sundaresan, N. R. et al. The deacetylase SIRT1 promotes membrane localization and activation of Akt and PDK1 during tumorigenesis and cardiac hypertrophy. Sci. Signal. 4, ra46 (2011).
Yang, W. L. et al. The E3 ligase TRAF6 regulates Akt ubiquitination and activation. Science 325, 1134–1138 (2009).
Chan, C. H. et al. The Skp2–SCF E3 ligase regulates Akt ubiquitination, glycolysis, herceptin sensitivity, and tumorigenesis. Cell 149, 1098–1111 (2012).
Essers, M. A. et al. FOXO transcription factor activation by oxidative stress mediated by the small GTPase Ral and JNK. EMBO J. 23, 4802–4812 (2004).
Wu, X. et al. Rac1 activation controls nuclear localization of β-catenin during canonical Wnt signaling. Cell 133, 340–353 (2008).
Mizukami, Y., Yoshioka, K., Morimoto, S. & Yoshida, K. A novel mechanism of JNK1 activation. Nuclear translocation and activation of JNK1 during ischemia and reperfusion. J. Biol. Chem. 272, 16657–16662 (1997).
Lin, K., Hsin, H., Libina, N. & Kenyon, C. Regulation of the Caenorhabditis elegans longevity protein DAF-16 by insulin/IGF-1 and germline signaling. Nature Genet. 28, 139–145 (2001).
Furuyama, T., Nakazawa, T., Nakano, I. & Mori, N. Identification of the differential distribution patterns of mRNAs and consensus binding sequences for mouse DAF-16 homologues. Biochem. J. 349, 629–634 (2000).
Ramaswamy, S., Nakamura, N., Sansal, I., Bergeron, L. & Sellers, W. R. A novel mechanism of gene regulation and tumor suppression by the transcription factor FKHR. Cancer Cell 2, 81–91 (2002).
Barr, F. G. Gene fusions involving PAX and FOX family members in alveolar rhabdomyosarcoma. Oncogene 20, 5736–5746 (2001).
Burgering, B. M. & Medema, R. H. Decisions on life and death: FOXO forkhead transcription factors are in command when PKB/Akt is off duty. J. Leukoc. Biol. 73, 689–701 (2003).
Lalmansingh, A. S., Karmakar, S., Jin, Y. & Nagaich, A. K. Multiple modes of chromatin remodeling by forkhead box proteins. Biochim. Biophys. Acta 1819, 707–715 (2012).
Hatta, M. & Cirillo, L. A. Chromatin opening and stable perturbation of core histone:DNA contacts by FoxO1. J. Biol. Chem. 282, 35583–35593 (2007).
Hatta, M., Liu, F. & Cirillo, L. A. Acetylation curtails nucleosome binding, not stable nucleosome remodeling, by FoxO1. Biochem. Biophys. Res. Commun. 379, 1005–1008 (2009).
van der Vos, K. E. & Coffer, P. J. FOXO-binding partners: it takes two to tango. Oncogene 27, 2289–2299 (2008).
You, H. et al. p53-dependent inhibition of FKHRL1 in response to DNA damage through protein kinase SGK1. Proc. Natl Acad. Sci. USA 101, 14057–14062 (2004).
Renault, V. M. et al. The pro-longevity gene FoxO3 is a direct target of the p53 tumor suppressor. Oncogene 30, 3207–3221 (2011).
Bouchard, C. et al. FoxO transcription factors suppress Myc-driven lymphomagenesis via direct activation of Arf. Genes Dev. 21, 2775–2787 (2007).
You, H., Yamamoto, K. & Mak, T. W. Regulation of transactivation-independent proapoptotic activity of p53 by FOXO3a. Proc. Natl Acad. Sci. USA 103, 9051–9056 (2006).
Kloet, D. E. & Burgering, B. M. The PKB/FOXO switch in aging and cancer. Biochim. Biophys. Acta 1813, 1926–1937 (2011).
Ferber, E. C. et al. FOXO3a regulates reactive oxygen metabolism by inhibiting mitochondrial gene expression. Cell Death Differ. 19, 968–979 (2012).
Jensen, K. S. et al. FoxO3A promotes metabolic adaptation to hypoxia by antagonizing Myc function. EMBO J. 30, 4554–4570 (2011).
Delpuech, O. et al. Induction of Mxi1-SR α by FOXO3a contributes to repression of Myc-dependent gene expression. Mol. Cell. Biol. 27, 4917–4930 (2007). Describes for the first time the molecular details as to how FOXO can antagonize MYC function.
Hoogeboom, D. & Burgering, B. M. Should I stay or should I go: β-catenin decides under stress. Biochim. Biophys. Acta 1796, 63–74 (2009).
Essers, M. A. et al. Functional interaction between β-catenin and FOXO in oxidative stress signaling. Science 308, 1181–1184 (2005). Shows the interaction of FOXO with β-catenin and so provides a novel link between WNT and insulin signalling that is regulated by oxidative stress. Provides biochemical rationale for the link between WNT–TCF and diabetes onset shown in reference 106.
Hoogeboom, D. et al. Interaction of FOXO with β-catenin inhibits β-catenin/T cell factor activity. J. Biol. Chem. 283, 9224–9230 (2008).
Almeida, M., Han, L., Martin-Millan, M., O'Brien, C. A. & Manolagas, S. C. Oxidative stress antagonizes Wnt signaling in osteoblast precursors by diverting β-catenin from T cell factor- to forkhead box O-mediated transcription. J. Biol. Chem. 282, 27298–27305 (2007).
Xie, X. W., Liu, J. X., Hu, B. & Xiao, W. Zebrafish foxo3b negatively regulates canonical Wnt signaling to affect early embryogenesis. PLoS ONE 6, e24469 (2011).
Kanazawa, A. et al. Association of the gene encoding wingless-type mammary tumor virus integration-site family member 5B (WNT5B) with type 2 diabetes. Am. J. Hum. Genet. 75, 832–843 (2004).
Grant, S. F. et al. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nature Genet. 38, 320–323 (2006). Describes a strong association between the classic WNT signalling transcription factor TFC4 and diabetes onset, providing the first in vivo evidence that insulin and WNT signalling may be linked through the regulation of TCF activity.
Mani, A. et al. LRP6 mutation in a family with early coronary disease and metabolic risk factors. Science 315, 1278–1282 (2007).
Liu, H. et al. Wnt signaling regulates hepatic metabolism. Sci. Signal. 4, ra6 (2011).
Hui, R. C. et al. The forkhead transcription factor FOXO3a increases phosphoinositide-3 kinase/Akt activity in drug-resistant leukemic cells through induction of PIK3CA expression. Mol. Cell. Biol. 28, 5886–5898 (2008).
Ide, T. et al. SREBPs suppress IRS-2-mediated insulin signalling in the liver. Nature Cell Biol. 6, 351–357 (2004).
Puig, O. & Tjian, R. Transcriptional feedback control of insulin receptor by dFOXO/FOXO1. Genes Dev. 19, 2435–2446 (2005).
Chen, C. C. et al. FoxOs inhibit mTORC1 and activate Akt by inducing the expression of Sestrin3 and Rictor. Dev. Cell 18, 592–604 (2010).
Kousteni, S. FoxO1, the transcriptional chief of staff of energy metabolism. Bone 50, 437–443 (2012).
Ren, H. et al. FoxO1 target Gpr17 activates AgRP neurons to regulate food intake. Cell 149, 1314–1326 (2012).
Lu, M. et al. Insulin regulates liver metabolism in vivo in the absence of hepatic Akt and Foxo1. Nature Med. 18, 388–395 (2012). Shows that the main function of PKB in insulin signalling in the liver is to control FOXO1 and reveals PKB-independent regulation of insulin-induced glucose metabolism.
Paradis, S. & Ruvkun, G. Caenorhabditis elegans Akt/PKB transduces insulin receptor-like signals from AGE-1 PI3 kinase to the DAF-16 transcription factor. Genes Dev. 12, 2488–2498 (1998).
Selkoe, D. J. Folding proteins in fatal ways. Nature 426, 900–904 (2003).
Morley, J. F., Brignull, H. R., Weyers, J. J. & Morimoto, R. I. The threshold for polyglutamine-expansion protein aggregation and cellular toxicity is dynamic and influenced by aging in Caenorhabditis elegans. Proc. Natl Acad. Sci. USA 99, 10417–10422 (2002).
Cohen, E., Bieschke, J., Perciavalle, R. M., Kelly, J. W. & Dillin, A. Opposing activities protect against age-onset proteotoxicity. Science 313, 1604–1610 (2006).
Cohen, E. et al. Temporal requirements of insulin/IGF-1 signaling for proteotoxicity protection. Aging Cell 9, 126–134 (2010).
Vilchez, D. et al. RPN-6 determines C. elegans longevity under proteotoxic stress conditions. Nature 489, 263–268 (2012).
Zhao, J. et al. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 6, 472–483 (2007).
Mammucari, C. et al. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 6, 458–471 (2007).
Mei, Y. et al. FOXO3a-dependent regulation of Pink1 (Park6) mediates survival signaling in response to cytokine deprivation. Proc. Natl Acad. Sci. USA 106, 5153–5158 (2009).
Sandri, M. et al. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell 117, 399–412 (2004). The first paper to suggest a role for FOXO in protein homeostasis other than regulating protein translation.
Bodine, S. C. et al. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science 294, 1704–1708 (2001).
Liu, H. Y. et al. Hepatic autophagy is suppressed in the presence of insulin resistance and hyperinsulinemia: inhibition of FoxO1-dependent expression of key autophagy genes by insulin. J. Biol. Chem. 284, 31484–31492 (2009).
Sengupta, A., Molkentin, J. D. & Yutzey, K. E. FoxO transcription factors promote autophagy in cardiomyocytes. J. Biol. Chem. 284, 28319–28331 (2009).
Zhao, Y. et al. Cytosolic FoxO1 is essential for the induction of autophagy and tumour suppressor activity. Nature Cell Biol. 12, 665–675 (2010).
Junger, M. A. et al. The Drosophila forkhead transcription factor FOXO mediates the reduction in cell number associated with reduced insulin signaling. J. Biol. 2, 20 (2003).
Puig, O., Marr, M. T., Ruhf, M. L. & Tjian, R. Control of cell number by Drosophila FOXO: downstream and feedback regulation of the insulin receptor pathway. Genes Dev. 17, 2006–2020 (2003). One of the first studies to focus on feedback control of signalling towards FOXO that is initiated by FOXO transcriptional activity.
Southgate, R. J. et al. FOXO1 regulates the expression of 4E-BP1 and inhibits mTOR signaling in mammalian skeletal muscle. J. Biol. Chem. 282, 21176–21186 (2007).
McColl, G. et al. Insulin-like signaling determines survival during stress via posttranscriptional mechanisms in C. elegans. Cell Metab. 12, 260–272 (2010).
Boehm, A. M. et al. FoxO is a critical regulator of stem cell maintenance in immortal Hydra. Proc. Natl Acad. Sci. USA 109, 19697–19702 (2012).
Miyamoto, K. et al. Foxo3a is essential for maintenance of the hematopoietic stem cell pool. Cell Stem Cell 1, 101–112 (2007).
Tothova, Z. et al. FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell 128, 325–339 (2007). Describes a role for FOXOs in adult stem cell maintenance.
Paik, J. H. et al. FoxOs cooperatively regulate diverse pathways governing neural stem cell homeostasis. Cell Stem Cell 5, 540–553 (2009).
Renault, V. M. et al. FoxO3 regulates neural stem cell homeostasis. Cell Stem Cell 5, 527–539 (2009).
Pan, G. & Thomson, J. A. Nanog and transcriptional networks in embryonic stem cell pluripotency. Cell Res. 17, 42–49 (2007).
Zhang, X. et al. FOXO1 is an essential regulator of pluripotency in human embryonic stem cells. Nature Cell Biol. 13, 1092–1099 (2011).
Becker, T. et al. FOXO-dependent regulation of innate immune homeostasis. Nature 463, 369–373 (2010).
Hedrick, S. M., Hess Michelini, R., Doedens, A. L., Goldrath, A. W. & Stone, E. L. FOXO transcription factors throughout T cell biology. Nature Rev. Immunol. 12, 649–661 (2012).
Dejean, A. S., Hedrick, S. M. & Kerdiles, Y. M. Highly specialized role of forkhead box O transcription factors in the immune system. Antioxid. Redox Signal. 14, 663–674 (2011).
Dengler, H. S. et al. Distinct functions for the transcription factor Foxo1 at various stages of B cell differentiation. Nature Immunol. 9, 1388–1398 (2008).
Kerdiles, Y. M. et al. Foxo1 links homing and survival of naive T cells by regulating L-selectin, CCR7 and interleukin 7 receptor. Nature Immunol. 10, 176–184 (2009).
Amin, R. H. & Schlissel, M. S. Foxo1 directly regulates the transcription of recombination-activating genes during B cell development. Nature Immunol. 9, 613–622 (2008).
Harada, Y. et al. Transcription factors Foxo3a and Foxo1 couple the E3 ligase Cbl-b to the induction of Foxp3 expression in induced regulatory T cells. J. Exp. Med. 207, 1381–1391 (2010).
Kerdiles, Y. M. et al. Foxo transcription factors control regulatory T cell development and function. Immunity 33, 890–904 (2010).
Ouyang, W. et al. Foxo proteins cooperatively control the differentiation of Foxp3+ regulatory T cells. Nature Immunol. 11, 618–627 (2010).
Ouyang, W. et al. Novel Foxo1-dependent transcriptional programs control TReg cell function. Nature 491, 554–559 (2012).
Litvak, V. et al. A FOXO3–IRF7 gene regulatory circuit limits inflammatory sequelae of antiviral responses. Nature 490, 421–425 (2012).
Seoane, J., Le, H. V., Shen, L., Anderson, S. A. & Massague, J. Integration of Smad and forkhead pathways in the control of neuroepithelial and glioblastoma cell proliferation. Cell 117, 211–223 (2004).
de Keizer, P. L. et al. Activation of forkhead box O transcription factors by oncogenic BRAF promotes p21cip1-dependent senescence. Cancer Res. 70, 8526–8536 (2010).
Naka, K. et al. TGF-β–FOXO signalling maintains leukaemia-initiating cells in chronic myeloid leukaemia. Nature 463, 676–680 (2010). Suggests that in cancer stem cells FOXOs have similar functions as in adult stem cells.
Sykes, S. M. et al. AKT/FOXO signaling enforces reversible differentiation blockade in myeloid leukemias. Cell 146, 697–708 (2011). Provides a rationale as to why the normal function of FOXO needs to be maintained during tumorigenesis and adds to the conclusion in reference 158 that FOXOs can also act to promote tumorigenesis.
Chakrabarty, A., Sanchez, V., Kuba, M. G., Rinehart, C. & Arteaga, C. L. Feedback upregulation of HER3 (ErbB3) expression and activity attenuates antitumor effect of PI3K inhibitors. Proc. Natl Acad. Sci. USA 109, 2718–2723 (2012).
Chandarlapaty, S. et al. AKT inhibition relieves feedback suppression of receptor tyrosine kinase expression and activity. Cancer Cell 19, 58–71 (2011).
Storz, P., Doppler, H., Copland, J. A., Simpson, K. J. & Toker, A. FOXO3a promotes tumor cell invasion through the induction of matrix metalloproteinases. Mol. Cell. Biol. 29, 4906–4917 (2009). Describes a pro-tumorigenic role for FOXO in driving metastasis, which is further supported by reference 159.
Tenbaum, S. P. et al. β-catenin confers resistance to PI3K and AKT inhibitors and subverts FOXO3a to promote metastasis in colon cancer. Nature Med. 18, 892–901 (2012).
van der Vos, K. E. & Coffer, P. J. The extending network of FOXO transcriptional target genes. Antioxid. Redox Signal. 14, 579–592 (2011).
Acknowledgements
The authors thank G. Kops and R. Martinez for critical reading of the manuscript and discussion. Research in the Burgering laboratory is funded by the Dutch Cancer Society (KWF), the Netherlands Organisation for Scientific Research (NWO) and the Centre of Biomedical Genetics (CBG). A.E. is supported by the Centre of Translational Molecular Medicine (CTMM) Research.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Glossary
- Homeostasis
-
The ability of an organism, or its constituent cells and organs, to establish and maintain an equilibrium that stabilizes its internal milieu and optimizes its ability to deal with moderate external changes.
- Dauer stage
-
An alternative developmental stage of nematode worms, including Caenorhabditis elegans, in which larva are adapted so that they can survive harsh conditions for an extended period of time.
- CBP–p300
-
A co-activator family composed of two closely related transcriptional co-activators. They regulate transcription by relaxing chromatin structures through histone acetylation, by recruiting the basal transcription machinery, including RNA polymerase II, and by acting as adaptor molecules.
- Methylation
-
The addition of a methyl group to a substrate. Protein methylation typically takes place on Arg or Lys residues, adding a maximum of two or three methyl groups, respectively.
- O-linked-D-N-acetylglucosamine
-
(O-GlcNAc). A monosaccharide derivative of glucose that is added as a post-translational modification on Ser and Thr residues.
- Cys oxidation
-
The thiol group of Cys residues can be easily oxidized, which can result in the formation of disulphide bonds with a Cys residue in the same or another protein.
- Epigenetic code
-
Hypothesized to be a defining code in every eukaryotic cell that consists of specific epigenetic modifications. It includes histone modifications defined by the histone code and additional epigenetic modifications such as DNA methylation.
- microRNAs
-
(miRNAs). Short RNAs found in eukaryotic cells that can post-transcriptionally regulate gene expression through binding to complementary sequences in target mRNAs. This usually results in translational repression or target degradation.
- Haploinsufficient
-
Haploinsufficiency occurs when a diploid organism only has a single functional copy of a gene, and the single functional copy of the gene does not produce enough of a gene product, which leads to an abnormal or diseased state.
- T cells
-
Lymphocytes that are essential components of the adaptive immune system and are mainly involved in cell-mediated immunity.
- PML bodies
-
(Promyelocytic leukaemia bodies). Spherical bodies that are found scattered throughout the nucleoplasm. PML bodies have been suggested to affect transcription regulation.
- Nucleolus
-
A non-membrane bound structure composed of proteins and nucleic acids found within the nucleus. The nucleolus is the location of rRNA transcription and assembly but can also capture and immobilize proteins.
- Pioneer factors
-
Although the precise definition is still debated, pioneer factors are thought to be transcription factors that can initially bind regulatory sequences, allowing binding of other factors by opening compacted chromatin, ultimately enabling transcriptional activation.
- Metabolic syndrome
-
A combination of medical disorders that, when occurring together, increase the risk of developing cardiovascular disease and diabetes.
- AGRP-neurons
-
(Agouti-related protein (AGRP)-expressing neurons). Hypothalamic neurons that express AGRP regulate food intake by stimulating feeding and inhibiting satiety.
- Autophagy
-
A tightly regulated catabolic process (also known as autophagocytosis) that involves the degradation of cellular components through the lysosomal machinery.
- Mitophagy
-
An autophagy pathway that selectively degrades mitochondria.
- Pluripotency
-
Pluripotency refers to a state of a cell, which has the potential to differentiate into any of the three germ layers (endoderm, mesoderm and ectoderm) and can give rise to any fetal or adult cell type.
- B cell
-
A lymphocyte and essential component of the adaptive immune system. The principal functions of B cells are to generate antibodies against antigens, act as antigen-presenting cells and eventually develop into memory B cells.
- Regulatory T cells
-
A subset of T cells that are also known as suppressor T cells. Their major role is to shut down T cell-mediated immunity towards the end of an immune reaction.
Rights and permissions
About this article
Cite this article
Eijkelenboom, A., Burgering, B. FOXOs: signalling integrators for homeostasis maintenance. Nat Rev Mol Cell Biol 14, 83–97 (2013). https://doi.org/10.1038/nrm3507
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrm3507
This article is cited by
-
FOXO transcription factors as mediators of stress adaptation
Nature Reviews Molecular Cell Biology (2024)
-
Emerging roles of mitochondrial functions and epigenetic changes in the modulation of stem cell fate
Cellular and Molecular Life Sciences (2024)
-
ERBB2-PTGS2 axis promotes intervertebral disc degeneration by regulating senescence of nucleus pulposus cells
BMC Musculoskeletal Disorders (2023)
-
The remoulding of dietary effects on the fecundity / longevity trade-off in a social insect
BMC Genomics (2023)
-
Diverse activity of miR-150 in Tumor development: shedding light on the potential mechanisms
Cancer Cell International (2023)