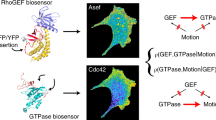
Abstract
Cellular signal transduction occurs in complex and redundant interaction networks, which are best understood by simultaneously monitoring the activation dynamics of multiple components. Recent advances in biosensor technology have made it possible to visualize and quantify the activation of multiple network nodes in the same living cell. The precision and scope of this approach has been greatly extended by novel computational approaches (referred to as computational multiplexing) that can reveal relationships between network nodes imaged in separate cells.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Olsen, J. V. et al. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127, 635–648 (2006).
Barrios-Rodiles, M. et al. High-throughput mapping of a dynamic signaling network in mammalian cells. Science 307, 1621–1625 (2005).
Margolin, A. A. et al. ARACNE: An algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinformatics 7 (Suppl. 1), S7 (2006).
Spencer, S. L., Gaudet, S., Albeck, J. G., Burke, J. M. & Sorger, P. K. Non-genetic origins of cell-to-cell variability in TRAIL-induced apoptosis. Nature 459, 428–432 (2009).
Colman-Lerner, A. et al. Regulated cell-to-cell variation in a cell-fate decision system. Nature 437, 699–706 (2005).
Cohen-Saidon, C., Cohen, A. A., Sigal, A., Liron, Y. & Alon, U. Dynamics and variability of ERK2 response to EGF in individual living cells. Mol. Cell 36, 885–893 (2009).
Sachs, K., Perez, O., Pe'er, D., Lauffenburger, D. A. & Nolan, G. P. Causal protein-signaling networks derived from multiparameter single-cell data. Science 308, 523–529 (2005).
Salmon, W. C., Adams, M. C. & Waterman-Storer, C. M. Dual-wavelength fluorescent speckle microscopy reveals coupling of microtubule and actin movements in migrating cells. J. Cell Biol. 158, 31–37 (2002).
Schaefer, A. W. et al. Coordination of actin filament and microtubule dynamics during neurite outgrowth. Dev. Cell 15, 146–162 (2008).
Hu, K., Ji, L., Applegate, K. T., Danuser, G. & Waterman-Storer, C. M. Differential transmission of actin motion within focal adhesions. Science 315, 111–115 (2007).
Taylor, M. J., Perrais, D. & Merrifield, C. J. A high precision survey of the molecular dynamics of mammalian clathrin-mediated endocytosis. PLoS Biol. 9, e1000604 (2011).
Harbeck, M. C. et al. Simultaneous optical measurements of cytosolic Ca2+ and cAMP in single cells. Sci. STKE 2006, PL6 (2006).
Wier, W. G., Rizzo, M. A., Raina, H. & Zacharia, J. A technique for simultaneous measurement of Ca2+, FRET fluorescence and force in intact mouse small arteries. J. Physiol. 586, 2437–2443 (2008).
Tengholm, A., Teruel, M. N. & Meyer, T. Single cell imaging of PI3K activity and glucose transporter insertion into the plasma membrane by dual color evanescent wave microscopy. Sci. STKE 2003, PL4 (2003).
Tanimura, A., Nezu, A., Morita, T., Turner, R. J. & Tojyo, Y. Fluorescent biosensor for quantitative real-time measurements of inositol 1,4,5-trisphosphate in single living cells. J. Biol. Chem. 279, 38095–38098 (2004).
Kitano, M., Nakaya, M., Nakamura, T., Nagata, S. & Matsuda, M. Imaging of Rab5 activity identifies essential regulators for phagosome maturation. Nature 453, 241–245 (2008).
VanEngelenburg, S. B. & Palmer, A. E. Fluorescent biosensors of protein function. Curr. Opin. Chem. Biol. 12, 60–65 (2008).
Newman, R. H., Fosbrink, M. D. & Zhang, J. Genetically encodable fluorescent biosensors for tracking signaling dynamics in living cells. Chem. Rev. 111, 3614–3666 (2011).
Hoppe, A. D. & Swanson, J. A. Cdc42, Rac1, and Rac2 display distinct patterns of activation during phagocytosis. Mol. Biol. Cell 15, 3509–3519 (2004).
Swanson, J. A. & Hoppe, A. D. The coordination of signaling during Fc receptor-mediated phagocytosis. J. Leukoc. Biol. 76, 1093–1103 (2004).
Vaughan, E. M., Miller, A. L., Yu, H. Y. & Bement, W. M. Control of local Rho GTPase crosstalk by Abr. Curr. Biol. 21, 270–277 (2011).
Machacek, M. et al. Coordination of Rho GTPase activities during cell protrusion. Nature 461, 99–103 (2009).
Ji, L., Lim, J. & Danuser, G. Fluctuations of intracellular forces during cell protrusion. Nature Cell Biol. 10, 1393–1400 (2008).
Tsukada, Y. et al. Quantification of local morphodynamics and local GTPase activity by edge evolution tracking. PLoS Comput. Biol. 4, e1000223 (2008).
Matz, M. V. et al. Fluorescent proteins from nonbioluminescent Anthozoa species. Nature Biotech. 17, 969–973 (1999).
Shaner, N. C. et al. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nature Biotech. 22, 1567–1572 (2004).
Shaner, N. C. et al. Improving the photostability of bright monomeric orange and red fluorescent proteins. Nature Methods 5, 545–551 (2008).
Piljic, A. & Schultz, C. Simultaneous recording of multiple cellular events by FRET. ACS Chem. Biol. 3, 156–160 (2008).
Ai, H. W., Hazelwood, K. L., Davidson, M. W. & Campbell, R. E. Fluorescent protein FRET pairs for ratiometric imaging of dual biosensors. Nature Methods 5, 401–403 (2008).
Niino, Y., Hotta, K. & Oka, K. Blue fluorescent cGMP sensor for multiparameter fluorescence imaging. PLoS ONE 5, e9164 (2010).
Niino, Y., Hotta, K. & Oka, K. Simultaneous live cell imaging using dual FRET sensors with a single excitation light. PLoS ONE 4, e6036 (2009).
Tomosugi, W. et al. An ultramarine fluorescent protein with increased photostability and pH insensitivity. Nature Methods 6, 351–353 (2009).
Jares-Erijman, E. A. & Jovin, T. M. FRET imaging. Nature Biotech. 21, 1387–1395 (2003).
Jares-Erijman, E. A. & Jovin, T. M. Imaging molecular interactions in living cells by FRET microscopy. Curr. Opin. Chem. Biol. 10, 409–416 (2006).
Wallrabe, H. & Periasamy, A. Imaging protein molecules using FRET and FLIM microscopy. Curr. Opin. Biotechnol. 16, 19–27 (2005).
Peyker, A., Rocks, O. & Bastiaens, P. I. Imaging activation of two Ras isoforms simultaneously in a single cell. Chembiochem 6, 78–85 (2005).
Murakoshi, H., Lee, S. J. & Yasuda, R. Highly sensitive and quantitative FRET–FLIM imaging in single dendritic spines using improved non-radiative YFP. Brain Cell Biol. 36, 31–42 (2008).
Ganesan, S., Ameer-Beg, S. M., Ng, T. T., Vojnovic, B. & Wouters, F. S. A dark yellow fluorescent protein (YFP)-based resonance energy-accepting chromoprotein (REACh) for Förster resonance energy transfer with GFP. Proc. Natl Acad. Sci. USA 103, 4089–4094 (2006).
Kwok, S. et al. Genetically encoded probe for fluorescence lifetime imaging of CaMKII activity. Biochem. Biophys. Res. Commun. 369, 519–525 (2008).
Kremers, G. J., van Munster, E. B., Goedhart, J. & Gadella, T. W. Jr. Quantitative lifetime unmixing of multiexponentially decaying fluorophores using single-frequency fluorescence lifetime imaging microscopy. Biophys. J. 95, 378–389 (2008).
Goedhart, J., Vermeer, J. E., Adjobo-Hermans, M. J., van Weeren, L. & Gadella, T. W. Jr. Sensitive detection of p65 homodimers using red-shifted and fluorescent protein-based FRET couples. PLoS ONE 2, e1011 (2007).
Shcherbo, D. et al. Practical and reliable FRET/FLIM pair of fluorescent proteins. BMC Biotechnol. 9, 24 (2009).
Padilla-Parra, S. et al. Quantitative comparison of different fluorescent protein couples for fast FRET-FLIM acquisition. Biophys. J. 97, 2368–2376 (2009).
Sapsford, K. E., Berti, L. & Medintz, I. L. Materials for fluorescence resonance energy transfer analysis: beyond traditional donor-acceptor combinations. Angew. Chem. Int. Ed. Engl. 45, 4562–4589 (2006).
Pertz, O. & Hahn, K. M. Designing biosensors for Rho family proteins--deciphering the dynamics of Rho family GTPase activation in living cells. J. Cell Sci. 117, 1313–1318 (2004).
Hinner, M. J. & Johnsson, K. How to obtain labeled proteins and what to do with them. Curr. Opin. Biotechnol. 21, 766–776 (2010).
Hu, C.-D. & Kerppola, T. K. Simultaneous visualization of multiple protein interactions in living cells using multicolor fluorescence complementation analysis. Nature Biotech. 21, 539–545 (2003).
Kodama, Y. & Wada, M. Simultaneous visualization of two protein complexes in a single plant cell using multicolor fluorescence complementation analysis. Plant Mol. Biol. 70, 211–217 (2009).
Vidi, P. A., Chemel, B. R., Hu, C. D. & Watts, V. J. Ligand-dependent oligomerization of dopamine D2 and adenosine A2A receptors in living neuronal cells. Mol. Pharmacol. 74, 544–551 (2008).
Grinberg, A. V., Hu, C. D. & Kerppola, T. K. Visualization of Myc/Max/Mad family dimers and the competition for dimerization in living cells. Mol. Cell Biol. 24, 4294–4308 (2004).
Michnick, S. W., Ear, P. H., Manderson, E. N., Remy, I. & Stefan, E. Universal strategies in research and drug discovery based on protein-fragment complementation assays. Nature Rev. Drug Discov. 6, 569–582 (2007).
Chu, J. et al. A novel far-red bimolecular fluorescence complementation system that allows for efficient visualization of protein interactions under physiological conditions. Biosens. Bioelectron. 25, 234–239 (2009).
Altman, D., Goswami, D., Hasson, T., Spudich, J. A. & Mayor, S. Precise positioning of myosin VI on endocytic vesicles in vivo. PLoS Biol. 5, e210 (2007).
Sharma, P. et al. Nanoscale organization of multiple GPI-anchored proteins in living cell membranes. Cell 116, 577–589 (2004).
Varma, R. & Mayor, S. GPI-anchored proteins are organized in submicron domains at the cell surface. Nature 394, 798–801 (1998).
Pertz, O., Hodgson, L., Klemke, R. L. & Hahn, K. M. Spatiotemporal dynamics of RhoA activity in migrating cells. Nature 440, 1069–1072 (2006).
Nalbant, P., Hodgson, L., Kraynov, V., Toutchkine, A. & Hahn, K. M. Activation of endogenous Cdc42 visualized in living cells. Science 305, 1615–1619 (2004).
Kraynov, V. S. et al. Localized Rac activation dynamics visualized in living cells. Science 290, 333–337 (2000).
Berlin, J. R., Bassani, J. W. & Bers, D. M. Intrinsic cytosolic calcium buffering properties of single rat cardiac myocytes. Biophys. J. 67, 1775–1787 (1994).
Helmchen, F., Imoto, K. & Sakmann, B. Ca2+ buffering and action potential-evoked Ca2+ signaling in dendrites of pyramidal neurons. Biophys. J. 70, 1069–1081 (1996).
Mochizuki, N. et al. Spatio-temporal images of growth-factor-induced activation of Ras and Rap1. Nature 411, 1065–1068 (2001).
Sawano, A., Takayama, S., Matsuda, M. & Miyawaki, A. Lateral propagation of EGF signaling after local stimulation is dependent on receptor density. Dev. Cell 3, 245–257 (2002).
Schultz, C., Schleifenbaum, A., Goedhart, J. & Gadella, T. W. Jr. Multiparameter imaging for the analysis of intracellular signaling. Chembiochem 6, 1323–1330 (2005).
Miyawaki, A. Visualization of the spatial and temporal dynamics of intracellular signaling. Dev. Cell 4, 295–305 (2003).
Bakal, C., Aach, J., Church, G. & Perrimon, N. Quantitative morphological signatures define local signaling networks regulating cell morphology. Science 316, 1753–1756 (2007).
Machacek, M. & Danuser, G. Morphodynamic profiling of protrusion phenotypes. Biophys. J. 90, 1439–1452 (2006).
Taylor, R. J. et al. Dynamic analysis of MAPK signaling using a high-throughput microfluidic single-cell imaging platform. Proc. Natl Acad. Sci. USA 106, 3758–3763 (2009).
Kiosses, W. B., Shattil, S. J., Pampori, N. & Schwartz, M. A. Rac recruits high-affinity integrin αvβ3 to lamellipodia in endothelial cell migration. Nature Cell Biol. 3, 316–320 (2001).
Wang, Y. et al. Visualizing the mechanical activation of Src. Nature 434, 1040–1045 (2005).
Levskaya, A., Weiner, O. D., Lim, W. A. & Voigt, C. A. Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature 461, 997–1001 (2009).
Wu, Y. I. et al. A genetically encoded photoactivatable Rac controls the motility of living cells. Nature 461, 104–108 (2009).
Tkachenko, E. et al. Protein kinase a governs a RhoA-RhoGDI-driven protrusion-retraction pacemaker in migrating cells. Nature Cell Biol. 13, 660–667 (2011).
Vilela, M. & Danuser, G. What's wrong with correlative experiments? Nature Cell Biol. 13, 1011 (2011).
Locasale, J. W. & Wolf-Yadlin, A. Maximum entropy reconstructions of dynamic signaling networks from quantitative proteomics data. PLoS ONE 4, e6522 (2009).
Perrin, B. E. et al. Gene networks inference using dynamic Bayesian networks. Bioinformatics 19 (Suppl. 2), ii138–ii148 (2003).
Sachs, K. et al. Learning signaling network structures with sparsely distributed data. J. Comput. Biol. 16, 201–212 (2009).
Gulyani, A. et al. A biosensor generated via high-throughput screening quantifies cell edge Src dynamics. Nature Chem. Biol. 7, 437–444 (2011).
Violin, J. D., Zhang, J., Tsien, R. Y. & Newton, A. C. A genetically encoded fluorescent reporter reveals oscillatory phosphorylation by protein kinase C. J. Cell Biol. 161, 899–909 (2003).
Takao, K. et al. Visualization of synaptic Ca2+/calmodulin-dependent protein kinase II activity in living neurons. J. Neurosci. 25, 3107–3112 (2005).
Acknowledgements
This work was funded by the US National Institutes of Health grants R01 GM90317 (to G.D. and K.H) and GM057464 (to K.H.) and the Cell Migration Consortium (U54 GM64346).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
41580_2011_BFnrm3212_MOESM1_ESM.pdf
Supplementary information S1 (Figure) | Spectra of fluorophores used in ratiometric FRET multiplex experiments. (PDF 566 kb)
Related links
Glossary
- Fluorescence polarization anisotropy
-
A technique that measures the rotational diffusion of fluorophores by measuring the difference in the polarization of excitation and emission light. Changes in fluorescence polarization anisotropy indicate changes in the rotational diffusion of molecules that are induced by their interactions with other molecules.
- Orthogonal wavelengths
-
Biosensor emission or excitation wavelengths that are sufficiently different to allow two fluorescent probes to be imaged separately in the same cell.
- Pairwise cross-correlation analysis
-
A technique that uses a mathematical framework to define whether the variation of one time-resolved image activity is coupled or independent of the variation of another time-resolved image activity.
- Quantum dots
-
Small semiconductor crystals that emit light of a longer wavelength on excitation with a shorter wavelength, akin to fluorophores.
- Ratiometric imaging
-
An imaging technique in which biosensors are designed so that the ratio of emission or excitation at two different wavelengths reflects the biological activity being measured. This ratio is independent of the biosensor's fluorescence intensity, so eliminates the effects of cell thickness, uneven biosensor distribution, uneven illumination and other factors.
- Spectral decomposition
-
A mathematical technique to separate the contribution of multiple fluorophore species to the image signal at a certain wavelength. This allows the separation of the signals of fluorophores with overlapping emission spectra.
- Stokes shift
-
The difference between the excitation and emission wavelengths of a fluorescent probe.
Rights and permissions
About this article
Cite this article
Welch, C., Elliott, H., Danuser, G. et al. Imaging the coordination of multiple signalling activities in living cells. Nat Rev Mol Cell Biol 12, 749–756 (2011). https://doi.org/10.1038/nrm3212
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrm3212
This article is cited by
-
Cell–extracellular matrix mechanotransduction in 3D
Nature Reviews Molecular Cell Biology (2023)
-
Förster resonance energy transfer biosensors for fluorescence and time-gated luminescence analysis of rac1 activity
Scientific Reports (2022)
-
Protein transfection via spherical nucleic acids
Nature Protocols (2022)
-
Phosphorylated Rho–GDP directly activates mTORC2 kinase towards AKT through dimerization with Ras–GTP to regulate cell migration
Nature Cell Biology (2019)
-
Intensiometric biosensors visualize the activity of multiple small GTPases in vivo
Nature Communications (2019)