Key Points
-
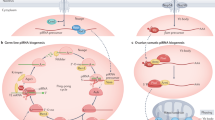
PIWI-interacting RNAs (piRNAs) are a distinct class of small non-coding RNAs that protect the integrity of the genome by silencing transposable elements in both germline and gonadal somatic cells. There are thousands of piRNA sequences in the genome and most reside in regions defined as piRNA clusters.
-
The pathways that drive the biogenesis of piRNAs are distinct from those that produce other small non-coding RNAs, including endogenous small interfering RNAs and microRNAs. piRNAs can arise from both a primary processing pathway and an amplifying, 'ping-pong' cycle that further refines the piRNA pool to ensure an effective defence against transposons.
-
piRNAs associate with PIWI proteins to form a piRNA-induced silencing complex (piRISC). Efficient piRNA-mediated silencing also requires association of several Tudor-domain proteins with PIWI proteins, as well as other non-Tudor-domain proteins.
-
There is emerging evidence that some piRNAs may also target protein-coding genes in both the germ line and the soma. In addition, piRNAs affect chromatin structure and transcription through effects on de novo methylation at loci containing transposable elements.
-
piRNAs localize to granular cytoplasmic bodies, where piRNA production and processing seems to take place. Although these show proximity to other mRNA processing bodies, the functional significance of this is not yet clear.
Abstract
PIWI-interacting RNAs (piRNAs) are a distinct class of small non-coding RNAs that form the piRNA-induced silencing complex (piRISC) in the germ line of many animal species. The piRISC protects the integrity of the genome from invasion by 'genomic parasites' — transposable elements — by silencing them. Owing to their limited expression in gonads and their sequence diversity, piRNAs have been the most mysterious class of small non-coding RNAs regulating RNA silencing. Now, much progress is being made into our understanding of their biogenesis and molecular functions, including the specific subcellular compartmentalization of the piRNA pathway in granular cytoplasmic bodies.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Ghildiyal, M. & Zamore, P. D. Small silencing RNAs: an expanding universe. Nature Rev. Genet. 10, 94–108 (2009).
Kim, N. V., Han, J. & Siomi M. C. Biogenesis of small RNAs in animals. Nature Rev. Mol. Cell Biol. 10, 126–139 (2009).
Malone, C. D. & Hannon, G. J. Small RNAs as guardians of the genome. Cell 136, 656–668 (2009).
Siomi, H. & Siomi, M. C. On the road to reading the RNA-interference code. Nature 457, 396–404 (2009).
Thomson, T. & Lin, H. The biogenesis and function of PIWI proteins and piRNAs: progress and prospect. Annu. Rev. Cell Dev. Biol. 25, 355–376 (2009).
Aravin, A. A. et al. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr. Biol. 11, 1017–1027 (2001).
Aravin, A. A. et al. The small RNA profile during Drosophila melanogaster development. Dev. Cell 5, 337–350 (2003).
Vagin, V. V., Sigova, A., Li, C., Seitz, H., Gvozdev, V. & Zamore, P. D. A distinct small RNA pathway silences selfish genetic elements in the germline. Science 313, 320–324 (2006). This paper hinted for the first time that Dicer endonuclease activity — which is essential for miRNA and siRNA biogenesis — might be dispensable for piRNA biogenesis in D. melanogaster.
Saito, K. et al. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev. 20, 2214–2222 (2006).
Aravin, A. et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature 442, 203–207 (2006).
Girard, A., Sachidanandam, R., Hannon, G. J. & Carmell, M. A. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 442, 199–202 (2006).
Grivna, S. T., Beyret, E., Wang, Z. & Lin, H. A novel class of small RNAs in the mouse spermatogenic cells. Genes Dev. 20, 1709–1714 (2006). References 10–12 describe the discovery of piRNAs in the mammalian male germ line. These piRNAs were not enriched in transposon-derived sequences and were later named 'pachytene piRNAs' because they are initially expressed at the pachytene stage of meiosis.
Houwing, S. et al. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell 129, 69–82 (2007).
Kazazian, H. H. Jr. Mobile elements: drivers of genome evolution. Science 303, 1626–1632 (2004).
Saito, K. & Siomi, M. C. Small RNA-mediated quiescence of transposable elements in animals. Dev. Cell 19, 687–697 (2010).
Vagin, V. V. et al. The RNA interference proteins and vasa locus are involved in the silencing of retrotransposons in the female germline of Drosophila melanogaster. RNA Biol. 1, 54–58 (2004).
Kalmykova, A. I., Klenov, M. S. & Gvozdev, V. A. Argonaute protein PIWI controls mobilization of retrotransposons in the Drosophila male germline. Nucleic Acids Res. 33, 2052–2059 (2005).
Savitsky, M., Kwon, D., Georgiev, P., Kalmykova, A. & Gvozdev, V. Telomere elongation is under the control of the RNAi-based mechanism in the Drosophila germline. Genes Dev. 20, 345–354 (2006).
Li, C. et al. Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell 137, 509–521 (2009).
Cox, D. N. et al. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 12, 3715–3727 (1998).
Lin, H. & Spradling, A. C. A novel group of pumilio mutations affects the asymmetric division of germline stem cells in the Drosophila ovary. Development 124, 2463–2476 (1997).
Schmidt, A. et al. Genetic and molecular characterization of sting, a gene involved in crystal formation and meiotic drive in the male germ line of Drosophila melanogaster. Genetics 151, 749–760 (1999).
Cox, D. N., Chao, A. & Lin, H. piwi encodes a nucleoplasmic factor whose activity modulates the number and division rate of germline stem cells. Development 127, 503–514 (2000).
Malone, C. D. et al. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell 137, 522–535 (2009).
Carmell, M. A. et al. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev. Cell 12, 503–514 (2007).
Aravin, A. A., Sachidanandam, R., Girard, A., Fejes-Toth, K. & Hannon, G. J. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science 316, 744–747 (2007).
Kuramochi-Miyagawa, S. et al. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev. 22, 908–917 (2008).
Kuramochi-Miyagawa, S. et al. Two mouse piwi-related genes: miwi and mili. Mech. Dev. 108, 121–133 (2001).
Aravin, A. A. et al. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol. Cell 31, 785–799 (2008). References 26, 27 and 29 identify the link between piRNA silencing of transposable elements and CpG methylation of their genomic sequences in the male germ line, which is probably mediated by the nuclear MIWI2 complex. As the piRNAs that potentially guide this DNA methylation are expressed early during mouse gametogenesis, they were named 'pre-pachytene piRNAs'.
Klattenhoff, C. et al. Drosophila rasiRNA pathway mutations disrupt embryonic axis specification through activation of an ATR/Chk2 DNA damage response. Dev. Cell 12, 45–55 (2007).
Brennecke, J. et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128, 1089–1103 (2007).
Gunawardane, L. S. et al. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science 315, 1587–1590 (2007). References 31 and 32 proposed the amplification loop — also known as the 'ping-pong model' — for piRNA biogenesis by revealing the specific features or 'signature' of piRNAs that are associated with PIWI proteins in D. melanogaster ovaries.
Nishida, K. M. et al. Gene silencing mechanisms mediated by Aubergine piRNA complexes in Drosophila male gonad. RNA 13, 1911–1922 (2007).
Saito, K. et al. Roles for the Yb body components Armitage and Yb in primary piRNA biogenesis in Drosophila. Genes Dev. 24, 2493–2498 (2010).
Pal-Bhadra, M. et al. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science 303, 669–672 (2004).
Brower-Toland, B. et al. Drosophila PIWI associates with chromatin and interacts directly with HP1a. Genes Dev. 21, 2300–2311 (2007).
Yin, H. & Lin, H. An epigenetic activation role of Piwi and a Piwi-associated piRNA in Drosophila melanogaster. Nature 450, 304–308 (2007).
Moshkovich, N. & Lei E. P. HP1 recruitment in the absence of argonaute proteins in Drosophila. PLoS Genet. 6, e1000880 (2010).
Ghildiyal, M. et al. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science 320, 1077–1081 (2008).
Kawamura, Y. et al. Drosophila endogenous small RNAs bind to Argonaute 2 in somatic cells. Nature 453, 793–797 (2008).
Tam, O. H. et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature 453, 534–538 (2008).
Watanabe, T. et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature 453, 539–543 (2008).
Li, M. A., Alls, J. D., Avancini, R. M., Koo, K. & Godt, D. The large Maf factor Traffic Jam controls gonad morphogenesis in Drosophila. Nature Cell Biol. 5, 994–1000 (2003).
Saito, K. et al. A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature 461, 1296–1299 (2009). References 19, 24 and 44 provided new insights into piRNAs that are specifically expressed in ovarian somatic cells, mainly by providing evidence that the origins of, and processing factors regulating, these somatic piRNAs are unique from those of germline piRNAs.
Rouget, C. et al. Maternal mRNA deadenylation and decay by the piRNA pathway in the early Drosophila embryo. Nature 467, 1128–1132 (2010).
Klattenhoff, C. et al. The Drosophila HP1 homolog Rhino is required for transposon silencing and piRNA production by dual-strand clusters. Cell 138, 1137–1149 (2009).
Aravin, A. A., Hannon, G. J. & Brennecke, J. The Piwi–piRNA pathway provides an adaptive defense in the transposon arms race. Science 318, 761–764 (2007).
Lau, N. C. et al. Characterization of the piRNA complex from rat testes. Science 313, 363–367 (2006).
Houwing, S., Berezikov, E. & Ketting, R. F. Zili is required for germ cell differentiation and meiosis in zebrafish. EMBO J. 27, 2702–2711 (2008).
Kawaoka, S. et al. The Bombyx ovary-derived cell line endogenously expresses PIWI/PIWI-interacting RNA complexes. RNA 15, 1258–1264 (2009).
Robine, N. et al. A broadly conserved pathway generates 3'UTR-directed primary piRNAs. Curr. Biol. 19, 2066–2076 (2009).
Mi, S. et al. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell 133, 116–127 (2008).
Haase, A. D. et al. Probing the initiation and effector phases of the somatic piRNA pathway in Drosophila. Genes Dev. 24, 2499–2504 (2010).
Olivieri, D., Sykora, M. M., Sachidanandam, R., Mechtler, K. & Brennecke, J. An in vivo RNAi assay identifies major genetic and cellular requirements for primary piRNA biogenesis in Drosophila. EMBO J. 29, 3301–3317 (2010).
Qi, H. et al. The Yb body, a major site for Piwi-associated RNA biogenesis and a gateway for Piwi expression and transport to the nucleus in somatic cells. J. Biol. Chem. 286, 3789–3797 (2011). References 34, 54 and 55 provided evidence that cytoplasmic Yb-bodies are the sites for primary piRNA biogenesis in D. melanogaster . Both biochemical and genetic approaches suggested a requirement for ARMI, FS(1)YB and other factors, such as ZUC, in the primary piRNA pathway.
Szakmary, A., Reedy, M., Qi, H. & Lin, H. The Yb protein defines a novel organelle and regulates male germline stem cell self-renewal in Drosophila melanogaster. J. Cell Biol. 185, 613–627 (2009).
Prud'homme, N., Gans, M., Masson, M., Terzian, C. & Bucheton, A. Flamenco, a gene controlling the gypsy retrovirus of Drosophila melanogaster. Genetics 139, 697–711 (1995).
Desset, S., Meignin, C., Dastugue, B. & Vaury, C. COM, a heterochromatic locus governing the control of independent endogenous retroviruses from Drosophila melanogaster. Genetics 164, 501–509 (2003).
Mével-Ninio, M., Pelisson, A., Kinder, J., Campos, A. R. & Bucheton, A. The flamenco locus controls the gypsy and ZAM retroviruses and is required for Drosophila oogenesis. Genetics 175, 1615–1624 (2007).
Desset, S., Buchon, N., Meignin, C., Coiffet, M. & Vaury, C. In Drosophila melanogaster the COM locus directs the somatic silencing of two retrotransposons through both Piwi-dependent and -independent pathways. PLoS ONE 3, e1526 (2008).
Nagao, A., Mituyama, T., Huang, H., Chen, D., Siomi, M. C. & Siomi, H. Biogenesis pathways of piRNAs loaded onto AGO3 in the Drosophila testis. RNA 16, 2503–2515 (2010).
Brennecke, J. et al. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science 322, 1387–1392 (2008).
Bourc'his, D. & Voinnet, O. A small-RNA perspective on gametogenesis, fertilization, and early zygotic development. Science 330, 617–622 (2010).
Vagin, V. V. Hannon, G. J. & Aravin, A. A. Arginine methylation as a molecular signature of the Piwi small RNA pathway. Cell Cycle 8, 4003–4004 (2009).
Siomi, M. C., Mannen, T. & Siomi, H. How does the royal family of Tudor rule the PIWI-interacting RNA pathway? Genes Dev. 24, 636–646 (2010).
Kirino, Y. et al. Arginine methylation of Piwi proteins catalysed by dPRMT5 is required for Ago3 and Aub stability. Nature Cell Biol. 11, 652–658 (2009).
Nishida, K. M. et al. Functional involvement of Tudor and dPRMT5 in the piRNA processing pathway in Drosophila germlines. EMBO J. 28, 3820–3831 (2009).
Vagin, V. V. et al. Proteomic analysis of murine Piwi proteins reveals a role for arginine methylation in specifying interaction with Tudor family members. Genes Dev. 23, 1749–1762 (2009).
Chen, C. et al. Mouse Piwi interactome identifies binding mechanism of Tdrkh Tudor domain to arginine methylated Miwi. Proc. Natl Acad. Sci. USA 106, 20336–20341 (2009).
Reuter, M. et al. Loss of the Mili-interacting Tudor domain-containing protein-1 activates transposons and alters the Mili-associated small RNA profile. Nature Struct. Mol. Biol. 16, 639–646 (2009).
Shoji, M. et al. The TDRD9-MIWI2 complex is essential for piRNA-mediated retrotransposon silencing in the mouse male germline. Dev. Cell 17, 775–787 (2009).
Vasileva, A., Tiedau, D., Firooznia, A., Müller-Reichert, T. & Jessberger, R. Tdrd6 is required for spermiogenesis, chromatoid body architecture, and regulation of miRNA expression. Curr. Biol. 19, 630–639 (2009).
Wang, J., Saxe, J. P., Tanaka, T., Chuma, S. & Lin, H. Mili interacts with tudor domain-containing protein 1 in regulating spermatogenesis. Curr. Biol. 19, 640–644 (2009).
Kirino, Y. et al. Arginine methylation of vasa protein is conserved across phyla. J. Biol. Chem. 285, 8148–8154 (2010). References 66–74 identify direct interactions between the N termini of PIWI proteins — which carry dimethylated arginine residues — and proteins with Tudor domains that recognize this modification. The authors show that the PIWI–Tudor protein interactions are crucial for the proper intracellular localization of the piRNA machinery and its silencing mechanism.
Patil, V. S. & Kai, T. Repression of retroelements in Drosophila germline via piRNA pathway by the tudor domain protein Tejas. Curr. Biol. 20, 724–730 (2010).
Boswell, R. E. & Mahowald, A. P. tudor, a gene required for assembly of the germ plasm in Drosophila melanogaster. Cell 43, 97–104 (1985).
Thomson, T. & Lasko, P. Drosophila tudor is essential for polar granule assembly and pole cell specification, but not for posterior patterning. Genesis 40, 164–170 (2004).
Thomson, T. & Lasko, P. Tudor and its domains: germ cell formation from a Tudor perspective. Cell Res. 15, 281–291 (2005).
Arkov, A. L., Wang, J. Y. S., Ramos, A. & Lehmann, R. The role of Tudor domains in germline development and polar granule architecture. Development 133, 4053–4062 (2006).
Liu, H. et al. Structural basis for methylarginine-dependent recognition of Aubergine by Tudor. Genes Dev. 24, 1876–1881 (2010).
Liu, K. et al. Structural basis for recognition of arginine methylated Piwi proteins by the extended Tudor domain. Proc. Natl Acad. Sci. USA 107, 18398–18403 (2010).
Lim, A. K & Kai, T. Unique germ-line organelle, nuage, functions to repress selfish genetic elements in Drosophila melanogaster. Proc. Natl Acad. Sci. USA 104, 6714–6719 (2007).
Frost, R. J. et al. MOV10L1 is necessary for protection of spermatocytes against retrotransposons by Piwi-interacting RNAs. Proc. Natl Acad. Sci. USA 107, 11847–11852 (2010).
Zheng, K. et al. Mouse MOV10L1 associates with Piwi proteins and is an essential component of the Piwi-interacting RNA (piRNA) pathway. Proc. Natl Acad. Sci. USA 107, 11841–11846 (2010).
Liang, L., Diehl-Jones, W. & Lasko, P. Localization of vasa protein to the Drosophila pole plasm is independent of its RNA-binding and helicase activities. Development 120, 1201–1211 (1994).
Toyooka, Y. et al. Expression and intracellular localization of mouse Vasa-homologue protein during germ cell development. Mech. Dev. 93, 139–149 (2000).
Kuramochi-Miyagawa, S. et al. MVH in piRNA processing and gene silencing of retrotransposons. Genes Dev. 24, 887–892 (2010).
Sengoku. T., Nureki. O., Nakamura. A., Kobayashi. S. & Yokoyama. S. Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa. Cell 125, 287–300 (2006).
Soper, S. F. et al. Mouse maelstrom, a component of nuage, is essential for spermatogenesis and transposon repression in meiosis. Dev. Cell 15, 285–297 (2008).
Costa, Y. et al. Mouse MAELSTROM: the link between meiotic silencing of unsynapsed chromatin and microRNA pathway? Hum. Mol. Genet. 15, 2324–2334 (2006).
Findley, S. D., Tamanaha, M., Clegg, N. J. & Ruohola-Baker, H. Maelstrom, a Drosophila spindle-class gene, encodes a protein that colocalizes with Vasa and RDE1/AGO1 homolog, Aubergine, in nuage. Development 130, 859–871 (2003).
Aravin, A. A. et al. Cytoplasmic compartmentalization of the fetal piRNA pathway in mice. PLoS Genet. 5, e1000764 (2009).
Ma, L. et al. GASZ is essential for male meiosis and suppression of retrotransposon expression in the male germline. PLoS Genet. 5, e1000635 (2009).
Anne, J. & Mechler, B. M. Valois, a component of the nuage and pole plasm, is involved in assembly of these structures, and binds to Tudor and the methyltransferase Capsuléen. Development 132, 2167–2177 (2005).
Kotaja, N., Lin, H., Parvinen, M. & Sassone-Corsi, P. Interplay of PIWI/Argonaute protein MIWI and kinesin KIF17b in chromatoid bodies of male germ cells. J. Cell Sci. 119, 2819–2825 (2006).
Onohara, Y., Fujiwara, T., Yasukochi, T., Himeno, M. & Yokota, S. Localization of mouse vasa homolog protein in chromatoid body and related nuage structures of mammalian spermatogenic cells during spermatogenesis. Histochem. Cell Biol. 133, 627–639 (2010). References 92, 95 and 96 revealed that, in mouse germ cells, components of piRNA machinery are localized in specific granular structures that were originally known as CBs or intermitochondrial cement.
Kojima, K. et al. Associations between PIWI proteins and TDRD1/MTR-1 are critical for integrated subcellular localization in murine male germ cells. Genes Cells 14, 1155–1165 (2009).
al-Mukhtar, K. A. & Webb, A. C. An ultrastructural study of primordial germ cells, oogonia and early oocytes in Xenopus laevis. J. Embryol. Exp. Morphol. 26, 195–217 (1971).
Eddy, E. M. Germ plasm and the differentiation of the germ cell line. Int. Rev. Cytol. 43, 229–280 (1975).
Illmensee, K. & Mahowald, A. P. Transplantation of posterior polar plasm in Drosophila. Induction of germ cells at the anterior pole of the egg. Proc. Natl Acad. Sci. USA 71, 1016–1020 (1974).
Kotaja, N. et al. The chromatoid body of male germ cells: similarity with processing bodies and presence of Dicer and microRNA pathway components. Proc. Natl Acad. Sci. USA 103, 2647–2652 (2006).
Acknowledgements
We thank H. Siomi, A. Nagao, A. Webster, N. Perkins and I. Olovnikov for comments on the manuscript. The work in the laboratory of M.C.S. is supported by Ministry of Education, Culture, Sports, Science and Technology (MEXT) grants to K.S. and M.C.S. M.C.S. is supported by the Core Research for Evolutional Science and Technology (CREST) programme of the Japan Science and Technology Agency (JST). The work in the laboratory of A.A.A. is supported by the US National Institutes of Health (grants DP2 OD007371A and R00 HD057233-02 to A.A.A.) and the Ellison Medical Foundation.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Glossary
- Morphogen
-
A signalling molecule that provides positional information to cells by forming a concentration gradient across the developmental field, and elicits multiple different cellular responses to specify cell fates in a concentration-dependent manner.
- Maternal–zygotic transition
-
A developmental switch during embryogenesis from control by maternally provided gene products to control by zygotically provided gene products. During this transition, embryonic transcription is initiated and many maternal products are degraded.
- Pachytene stage
-
A stage in meiotic prophase when homologous chromosomes are completely paired and chromosomal crossover occurs.
- Phasing
-
An endonucleolytic processing pattern with an equal periodicity. Small interfering RNAs are endonucleolytically processed (or 'diced') by Dicer from individual precursors at certain intervals to produce a typical size of ∼21 nucleotides. Dicer acts as a 'ruler' during processing.
- Tudor domain
-
A conserved protein motif typically consisting of ∼50 amino-acid residues. It is commonly found in proteins, such as RNA-binding proteins. Some Tudor domains specifically recognize and bind symmetrically dimethylated arginines.
- Hypervariable antibody repertoire
-
The antibody specificities that can potentially be produced by an individual. A huge repertoire of different antibodies is generated in a single individual.
- Germline granule
-
A specialized electron-dense structure that comprises particular RNAs and proteins that are indispensable for germline development. Vasa, an RNA helicase, is a well-characterized component of germline granules.
- High mobility group box
-
(HMG-box). A protein domain involved in DNA binding that comprises ∼80 amino-acid residues that form three α-helices.
Rights and permissions
About this article
Cite this article
Siomi, M., Sato, K., Pezic, D. et al. PIWI-interacting small RNAs: the vanguard of genome defence. Nat Rev Mol Cell Biol 12, 246–258 (2011). https://doi.org/10.1038/nrm3089
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrm3089
This article is cited by
-
Epigenetic modifications in the development of bronchopulmonary dysplasia: a review
Pediatric Research (2024)
-
Small RNAs, spermatogenesis, and male infertility: a decade of retrospect
Reproductive Biology and Endocrinology (2023)
-
Reference genomes of channel catfish and blue catfish reveal multiple pericentric chromosome inversions
BMC Biology (2023)
-
Nanoparticle carriers enhance RNA stability and uptake efficiency and prolong the protection against Rhizoctonia solani
Phytopathology Research (2023)
-
Regulation of Miwi-mediated mRNA stabilization by Ck137956/Tssa is essential for male fertility
BMC Biology (2023)