Key Points
-
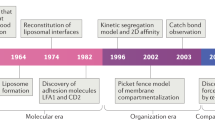
In juxtacrine signalling, surfaces of interacting cells come into direct contact and receptor–ligand recognition at this interface triggers intracellular signalling. Interactions involve multiple adhesion and signalling molecules, the collective behaviour of which regulates signal transduction.
-
The coordinated organization of cell membrane receptors into micrometre-scale patterns is emerging as a broadly significant theme of intercellular signalling, as exemplified by immunological synapses. Recent evidence suggests that this dynamic spatial organization has an active role in regulating the signalling state of individual molecular components and thus can alter long-term cell activation.
-
The physical mechanisms that establish and regulate the spatial organization of signalling molecules are equally as important as the chemical reactions themselves and both the cell membrane and the actin cytoskeleton have key roles.
-
The role of spatial organization is best appreciated by experiments that physically alter the organization of cell surface molecules at the interface between two cells. As a result, new experimental strategies have emerged to manipulate the spatial organization of molecules inside living cells.
Abstract
The coordinated organization of cell membrane receptors into diverse micrometre-scale spatial patterns is emerging as an important theme of intercellular signalling, as exemplified by immunological synapses. Key characteristics of these patterns are that they transcend direct protein–protein interactions, emerge transiently and modulate signal transduction. Such cooperativity over multiple length scales presents new and intriguing challenges for the study and ultimate understanding of cellular signalling. As a result, new experimental strategies have emerged to manipulate the spatial organization of molecules inside living cells. The resulting spatial mutations yield insights into the interweaving of the spatial, mechanical and chemical aspects of intercellular signalling.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Groves, J. T. Molecular organization and signal transduction at intermembrane junctions. Angew. Chem. Int. Ed. Engl. 44, 3524–3538 (2005).
Reich, Z. et al. Ligand-specific oligomerization of T-cell receptor molecules. Nature 387, 617–620 (1997).
Bray, D., Levin, M. D. & Morton-Firth, C. J. Receptor clustering as a cellular mechanism to control sensitivity. Nature 393, 85–88 (1998).
Maheshwari, G., Brown, G., Lauffenburger, D. A., Wells, A. & Griffith, L. G. Cell adhesion and motility depend on nanoscale RGD clustering. J. Cell Sci. 113, 1677–1686 (2000).
Grakoui, A. et al. The immunological synapse: a molecular machine controlling T cell activation. Science 285, 221–227 (1999).
Delon, J. & Germain, R. N. Information transfer at the immunological synapse. Curr. Biol. 10, R923–R933 (2000).
Davis, M. M. et al. Dynamics of cell surface molecules during T cell recognition. Annu. Rev. Biochem. 72, 717–742 (2003).
Davis, D. M. & Dustin, M. L. What is the importance of the immunological synapse? Trends Immunol. 25, 323–327 (2004).
Singleton, K. L. et al. Spatiotemporal patterning during T cell activation is highly diverse. Sci. Signal. 2, ra15 (2009).
van Der Merwe, P. A. & Davis, S. J. Immunology. The immunological synapse — a multitasking system. Science 295, 1479–1480 (2002).
Huppa, J. B. & Davis, M. M. T-cell-antigen recognition and the immunological synapse. Nature Rev. Immunol. 3, 973–983 (2003).
Dustin, M. L. & Colman, D. R. Neural and immunological synaptic relations. Science 298, 785–789 (2002).
Friedl, P., den Boer, A. T. & Gunzer, M. Tuning immune responses: diversity and adaptation of the immunological synapse. Nature Rev. Immunol. 5, 532–545 (2005).
Kao, H., Lin, J., Littman, D. R., Shaw, A. S. & Allen, P. M. Regulated movement of CD4 in and out of the immunological synapse. J. Immunol. 181, 8248–8257 (2008).
Cemerski, S. et al. The stimulatory potency of T cell antigens is influenced by the formation of the immunological synapse. Immunity 26, 345–355 (2007).
Cemerski, S. et al. The balance between T cell receptor signaling and degradation at the center of the immunological synapse is determined by antigen quality. Immunity 29, 414–422 (2008). Reports that the centre of the immunological synapse can be a site of sustained signalling for low doses of, or weak, antigens.
Mossman, K. D., Campi, G., Groves, J. T. & Dustin, M. L. Altered TCR signaling from geometrically repatterned immunological synapses. Science 310, 1191–1193 (2005). TCRs were physically restricted in the periphery of the synapse and their radial localization was shown to regulate their signalling.
Sims, T. N. et al. Opposing effects of PKCτ and WASp on symmetry breaking and relocation of the immunological synapse. Cell 129, 773–785 (2007).
Dustin, M. L. Cell adhesion molecules and actin cytoskeleton at immune synapses and kinapses. Curr. Opin. Cell Biol. 19, 529–533 (2007).
Paul, W. E. & Seder, R. A. Lymphocyte-responses and cytokines. Cell 76, 241–251 (1994).
Varma, R., Campi, G., Yokosuka, T., Saito, T. & Dustin, M. L. T cell receptor-proximal signals are sustained in peripheral microclusters and terminated in the central supramolecular activation cluster. Immunity 25, 117–127 (2006).
Monks, C. R., Freiberg, B. A., Kupfer, H., Sciaky, N. & Kupfer, A. Three-dimensional segregation of supramolecular activation clusters in T cells. Nature 395, 82–86 (1998).
Lee, K. H. et al. The immunological synapse balances T cell receptor signaling and degradation. Science 302, 1218–1222 (2003). Combines experiments and computer modelling to reveal the dual capacity of the immunological synapse to act as an enhancer and a downregulator of TCR signalling.
Dustin, M. L. T-cell activation through immunological synapses and kinapses. Immunol. Rev. 221, 77–89 (2008).
Gomez, T. S. & Billadeau, D. D. T cell activation and the cytoskeleton: you can't have one without the other. Adv. Immunol. 97, 1–64 (2008).
Orange, J. S. Formation and function of the lytic NK-cell immunological synapse. Nature Rev. Immunol. 8, 713–725 (2008).
Yokosuka, T. & Saito, T. Dynamic regulation of T-cell costimulation through TCR-CD28 microclusters. Immunol. Rev. 229, 27–40 (2009).
Hallman, E., Burack, W. R., Shaw, A. S., Dustin, M. L. & Allen, P. M. Immature CD4+CD8+ thymocytes form a multifocal immunological synapse with sustained tyrosine phosphorylation. Immunity 16, 839–848 (2002).
Davis, D. M. et al. The human natural killer cell immune synapse. Proc. Natl Acad. Sci. USA 96, 15062–15067 (1999).
Almeida, C. R. & Davis, D. M. Segregation of HLA-C from ICAM-1 at NK cell immune synapses is controlled by its cell surface density. J. Immunol. 177, 6904–6910 (2006).
Culley, F. J. et al. Natural killer cell signal integration balances synapse symmetry and migration. PLoS Biol. 7, e1000159 (2009).
Masilamani, M., Nguyen, C., Kabat, J., Borrego, F. & Coligan, J. E. CD94/NKG2A inhibits NK cell activation by disrupting the actin network at the immunological synapse. J. Immunol. 177, 3590–3596 (2006).
Doh, J. & Irvine, D. J. Immunological synapse arrays: patterned protein surfaces that modulate immunological synapse structure formation in T cells. Proc. Natl Acad. Sci. USA 103, 5700–5705 (2006). Developed a surface recreating canonical, multifocal and inverted synapses, and showed that some downstream T cell signalling pathways are altered by the new geometries.
Shen, K., Thomas, V. K., Dustin, M. L. & Kam, L. C. Micropatterning of costimulatory ligands enhances CD4+ T cell function. Proc. Natl Acad. Sci. USA 105, 7791–7796 (2008). Reveals that segregation, colocalization and relative radial juxtaposition alter how CD28 co-stimulates TCR signalling.
Yokosuka, T. et al. Spatiotemporal regulation of T cell costimulation by TCR-CD28 microclusters and protein kinase Cτ translocation. Immunity 29, 589–601 (2008).
Huppa, J. B. et al. TCR-peptide-MHC interactions in situ show accelerated kinetics and increased affinity. Nature. 463, 963–967 (2010).
Weikl, T. R. & Lipowsky, R. Pattern formation during T-cell adhesion. Biophys. J. 87, 3665–3678 (2004).
Altan-Bonnet, G. & Germain, R. N. Modeling T cell antigen discrimination based on feedback control of digital ERK responses. Plos Biol. 3, 1925–1938 (2005).
Wylie, D. C., Das, J. & Chakraborty, A. K. Sensitivity of T cells to antigen and antagonism emerges from differential regulation of the same molecular signaling module. Proc. Natl Acad. Sci. USA 104, 5533–5538 (2007).
Feinerman, O., Germain, R. N. & Altan-Bonnet, G. Quantitative challenges in understanding ligand discrimination by αβ T cells. Mol. Immunol. 45, 619–631 (2008).
Chakraborty, A. K. & Das, J. Pairing computation with experimentation: a powerful coupling for understanding T cell signalling. Nature Rev. Immunol. 10, 59–71 (2010).
Kastrup, C. J., Runyon, M. K., Shen, F. & Ismagilov, R. F. Modular chemical mechanism predicts spatiotemporal dynamics of initiation in the complex network of hemostasis. Proc. Natl Acad. Sci. USA 103, 15747–15752 (2006).
Kastrup, C. J., Shen, F., Runyon, M. K. & Ismagilov, R. F. Characterization of the threshold response of initiation of blood clotting to stimulus patch size. Biophys. J. 93, 2969–2977 (2007).
Veatch, S. L. et al. Critical fluctuations in plasma membrane vesicles. ACS Chem. Biol. 3, 287–293 (2008).
Lingwood, D. & Simons, K. Lipid rafts as a membrane-organizing principle. Science 327, 46–50 (2010).
Dustin, M. L., Ferguson, L. M., Chan, P. Y., Springer, T. A. & Golan, D. E. Visualization of CD2 interaction with LFA-3 and determination of the two-dimensional dissociation constant for adhesion receptors in a contact area. J. Cell Biol. 132, 465–474 (1996).
Groves, J. T. Bending mechanics and molecular organization in biological membranes. Annu. Rev. Phys. Chem. 58, 697–717 (2007).
Dustin, M. L. Adhesive bond dynamics in contacts between T lymphocytes and glass-supported planar bilayers reconstituted with the immunoglobulin-related adhesion molecule CD58. J. Biol. Chem. 272, 15782–15788 (1997).
Qi, S. Y., Groves, J. T. & Chakraborty, A. K. Synaptic pattern formation during cellular recognition. Proc. Natl Acad. Sci. USA 98, 6548–6553 (2001).
Weikl, T. R., Asfaw, M., Krobath, H., Rozycki, B. & Lipowsky, R. Adhesion of membranes via receptor-ligand complexes: domain formation, binding cooperativity, and active processes. Soft Matter 5, 3213–3224 (2009).
Burroughs, N. J., Lazic, Z. & van der Merwe, P. A. Ligand detection and discrimination by spatial relocalization: a kinase-phosphatase segregation model of TCR activation. Biophys. J. 91, 1619–1629 (2006).
Milstein, O. et al. Nanoscale increases in CD2-CD48-mediated intermembrane spacing decrease adhesion and reorganize the immunological synapse. J. Biol. Chem. 283, 34414–34422 (2008).
Kaizuka, Y., Douglass, A. D., Vardhana, S., Dustin, M. L. & Vale, R. D. The coreceptor CD2 uses plasma membrane microdomains to transduce signals in T cells. J. Cell Biol. 185, 521–534 (2009).
Choudhuri, K., Wiseman, D., Brown, M. H., Gould, K. & van der Merwe, P. A. T-cell receptor triggering is critically dependent on the dimensions of its peptide-MHC ligand. Nature 436, 578–582 (2005).
Billadeau, D. D., Nolz, J. C. & Gomez, T. S. Regulation of T-cell activation by the cytoskeleton. Nature Rev. Immunol. 7, 131–143 (2007).
Kaizuka, Y., Douglass, A. D., Varma, R., Dustin, M. L. & Vale, R. D. Mechanisms for segregating T cell receptor and adhesion molecules during immunological synapse formation in Jurkat T cells. Proc. Natl Acad. Sci. USA 104, 20296–20301 (2007).
DeMond, A. L., Mossman, K. D., Starr, T., Dustin, M. L. & Groves, J. T. T cell receptor microcluster transport through molecular mazes reveals mechanism of translocation. Biophys. J. 94, 3286–3292 (2008).
Burkhardt, J. K., Carrizosa, E. & Shaffer, M. H. The actin cytoskeleton in T cell activation. Annu. Rev. Immunol. 26, 233–259 (2008).
Ponti, A., Machacek, M., Gupton, S. L., Waterman-Storer, C. M. & Danuser, G. Two distinct actin networks drive the protrusion of migrating cells. Science 305, 1782–1786 (2004).
Chhabra, E. S. & Higgs, H. N. The many faces of actin: matching assembly factors with cellular structures. Nature Cell Biol. 9, 1110–1121 (2007).
Koretzky, G. A. & Myung, P. S. Positive and negative regulation of T-cell activation by adaptor proteins. Nature Rev. Immunol. 1, 95–107 (2001).
Smith, A. et al. A talin-dependent LFA-1 focal zone is formed by rapidly migrating T lymphocytes. J. Cell Biol. 170, 141–151 (2005).
Hartman, N. C., Nye, J. A. & Groves, J. T. Cluster size regulates protein sorting in the immunological synapse. Proc. Natl Acad. Sci. USA 106, 12729–12734 (2009). Proposes that frictional coupling to the actin cytoskeleton is a mechanism that controls the spatial patterning of membrane proteins.
Bunnell, S. C. et al. T cell receptor ligation induces the formation of dynamically regulated signaling assemblies. J. Cell Biol. 158, 1263–1275 (2002).
Nguyen, K., Sylvain, N. R. & Bunnell, S. C. T cell costimulation via the integrin VLA-4 inhibits the actin-dependent centralization of signaling microclusters containing the adaptor SLP-76. Immunity 28, 810–821 (2008).
Warren, C. M. & Landgraf, R. Signaling through ERBB receptors: multiple layers of diversity and control. Cell. Signal. 18, 923–933 (2006).
Tian, T. et al. Plasma membrane nanoswitches generate high-fidelity Ras signal transduction. Nature Cell Biol. 9, 905–914 (2007).
Salaita, K. et al. Restriction of receptor movement alters cellular response: physical force sensing by EphA2. Science 327, 1380–1385 (2010).
Greenfield, D. et al. Self-organization of the Escherichia coli chemotaxis network imaged with super-resolution light microscopy. Plos Biol. 7, e1000137 (2009).
Gestwicki, J. E. & Kiessling, L. L. Inter-receptor communication through arrays of bacterial chemoreceptors. Nature 415, 81–84 (2002).
Sourjik, V. & Berg, H. C. Functional interactions between receptors in bacterial chemotaxis. Nature 428, 437–441 (2004).
Lillemeier, B. F. et al. TCR and LAT are expressed on separate protein islands on T cell membranes and concatenate during activation. Nature Immunol. 11, 90–96 (2010).
Pautot, S., Lee, H., Isacoff, E. Y. & Groves, J. T. Neuronal synapse interaction reconstituted between live cells and supported lipid bilayers. Nature Chem. Biol. 1, 283–289 (2005).
Sackmann, E. Supported membranes: scientific and practical applications. Science 271, 43–48 (1996).
Groves, J. T. & Dustin, M. L. Supported planar bilayers in studies on immune cell adhesion and communication. J. Immunol. Methods 278, 19–32 (2003).
Groves, J. T. Spatial mutation of the T cell immunological synapse. Curr. Opin. Chem. Biol. 10, 544–550 (2006).
Nye, J. A. & Groves, J. T. Kinetic control of histidine-tagged protein surface density on supported lipid bilayers. Langmuir 24, 4145–4149 (2008).
Groves, J. T., Ulman, N. & Boxer, S. G. Micropatterning fluid lipid bilayers on solid supports. Science 275, 651–653 (1997).
Falconnet, D., Csucs, G., Grandin, H. M. & Textor, M. Surface engineering approaches to micropattern surfaces for cell-based assays. Biomaterials 27, 3044–3063 (2006).
Whitesides, G. M., Ostuni, E., Takayama, S., Jiang, X. & Ingber, D. E. Soft lithography in biology and biochemistry. Annu. Rev. Biomed. Eng. 3, 335–373 (2001).
Mossman, K. & Groves, J. Micropatterned supported membranes as tools for quantitative studies of the immunological synapse. Chem. Soc. Rev. 36, 46–54 (2007).
Nie, Z. & Kumacheva, E. Patterning surfaces with functional polymers. Nature Mater. 7, 277–290 (2008).
Sia, S. K. & Whitesides, G. M. Microfluidic devices fabricated in poly(dimethylsiloxane) for biological studies. Electrophoresis 24, 3563–3576 (2003).
Delamarche, E., Juncker, D. & Schmid, H. Microfluidics for processing surfaces and miniaturizing biological assays. Adv Mater. 17, 2911–2933 (2005).
Irvine, D. J., Doh, J. & Huang, B. Patterned surfaces as tools to study ligand recognition and synapse formation by T cells. Curr. Op. Immunol. 19, 463–469 (2007).
Ruiz, S. A. & Chen, C. S. Microcontact printing: a tool to pattern. Soft Matter 3, 168–177 (2007).
Salaita, K., Wang, Y. & Mirkin, C. A. Applications of dip-pen nanolithography. Nature Nanotechnol. 2, 145–155 (2007).
Girard, P. P., Cavalcanti-Adam, E. A., Kemkemer, R. & Spatz, J. P. Cellular chemomechanics at interfaces: sensing, integration and response. Soft Matter 3, 307–326 (2007).
Sackmann, E. & Tanaka, M. Supported membranes on soft polymer cushions: fabrication, characterization and applications. Trends Biotechnol. 18, 58–64 (2000).
Tanaka, M. & Sackmann, E. Polymer-supported membranes as models of the cell surface. Nature 437, 656–663 (2005).
Wu, M., Holowka, D., Craighead, H. G. & Baird, B. Visualization of plasma membrane compartmentalization with patterned lipid bilayers. Proc. Natl Acad. Sci. USA 101, 13798–13803 (2004).
Chen, C. S., Mrksich, M., Huang, S., Whitesides, G. M. & Ingber, D. E. Geometric control of cell life and death. Science 276, 1425–1428 (1997).
Arnold, M. et al. Activation of integrin function by nanopatterned adhesive interfaces. Chemphyschem. 5, 383–388 (2004).
Geiger, B., Spatz, J. P. & Bershadsky, A. D. Environmental sensing through focal adhesions. Nature Rev. Mol. Cell Biol. 10, 21–33 (2009).
Torres, A. J., Wu, M., Holowka, D. & Baird, B. Nanobiotechnology and cell biology: micro- and nanofabricated surfaces to investigate receptor-mediated signaling. Annu. Rev. Biophys. 37, 265–288 (2008).
Schwarzenbacher, M. et al. Micropatterning for quantitative analysis of protein-protein interactions in living cells. Nature Methods 5, 1053–1060 (2008).
Airan, R. D., Thompson, K. R., Fenno, L. E., Bernstein, H. & Deisseroth, K. Temporally precise in vivo control of intracellular signalling. Nature 458, 1025–1029 (2009).
Wu, Y. I. et al. A genetically encoded photoactivatable Rac controls the motility of living cells. Nature 461, 104–108 (2009).
Levskaya, A., Weiner, O. D., Lim, W. A. & Voigt, C. A. Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature 461, 997–1001 (2009).
Gorostiza, P. & Isacoff, E. Y. Optical switches for remote and noninvasive control of cell signaling. Science 322, 395–399 (2008).
Szobota, S. et al. Remote control of neuronal activity with a light-gated glutamate receptor. Neuron. 54, 535–545 (2007).
DeMond, A. L., Starr, T., Dustin, M. L. & Groves, J. T. Control of antigen presentation with a photoreleasable agonist peptide. J. Am. Chem. Soc. 128, 15354–15355 (2006).
Huse, M. et al. Spatial and temporal dynamics of T cell receptor signaling with a photoactivatable agonist. Immunity 27, 76–88 (2007).
Guo, Y. M. et al. Imaging dynamic cell-cell junctional coupling in vivo using Trojan-LAMP. Nature Methods 5, 835–841 (2008).
Meyer, T. & Teruel, M. N. Fluorescence imaging of signaling networks. Trends Cell Biol. 13, 101–106 (2003).
Sabouri-Ghomi, M., Wu, Y., Hahn, K. & Danuser, G. Visualizing and quantifying adhesive signals. Curr. Opin. Cell Biol. 20, 541–550 (2008).
Takayama, S. et al. Patterning cells and their environments using multiple laminar fluid flows in capillary networks. Proc. Natl Acad. Sci. USA 96, 5545–5548 (1999).
Acknowledgements
The hybrid live cell–supported membrane component of this work was supported by the Director, Office of Science, Office of Basic Energy Sciences, Chemical Sciences, Geosciences and Biosciences Division of the U.S. Department of Energy under contract number DE-AC02-05CH11231. General support for this project was also provided by a National Science Foundation CAREER award MCB-0448614 to J.T.G.
Author information
Authors and Affiliations
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Glossary
- Cytokine
-
A member of a large family of immunomodulating secreted proteins that interact with cellular receptors. Cytokine production results in the activation of an intracellular signalling cascade that commonly regulates processes such as inflammation.
- Miscibility phase separation
-
The partitioning of lipid components (in the context of membranes) into domains that have different chemical compositions and physical properties.
- LAT
-
(Linker for activation of T cells). A transmembrane protein that on TCR activation becomes rapidly phosphorylated and binds multiple adaptor molecules and indirectly recruits others.
- hsPALM
-
(High speed photoactivated localization microscopy). A fluorescence imaging technique in which sequential activation, localization and bleaching of fluorescent reporter proteins yields an image with a resolution of a few tens of nanometers, well below the diffraction limit.
- dcFFCS
-
(Dual colour fluorescence cross-correlation spectroscopy). A technique that analyses the dynamics and association of two different diffusing fluorescent proteins.
- Chemotaxis
-
Directed cell movement according to chemical stimuli.
- Liposome or proteoliposome
-
A vesicle made of lipid bilayer in an aqueous environment. Membrane proteins can be incorporated in the bilayer.
- Photo-switchable molecule
-
A molecule with a functionality (ligand binding, conformational change or absorption spectrum) that is controlled by light and in some cases can be toggled on and off.
- Uncaging
-
The light-controlled release of a functional group that hides (cages) another functional group.
Rights and permissions
About this article
Cite this article
Manz, B., Groves, J. Spatial organization and signal transduction at intercellular junctions. Nat Rev Mol Cell Biol 11, 342–352 (2010). https://doi.org/10.1038/nrm2883
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrm2883
This article is cited by
-
Adherens junctions organize size-selective proteolytic hotspots critical for Notch signalling
Nature Cell Biology (2022)
-
Dual Biomembrane Force Probe enables single-cell mechanical analysis of signal crosstalk between multiple molecular species
Scientific Reports (2017)
-
Human NK cell development requires CD56-mediated motility and formation of the developmental synapse
Nature Communications (2016)
-
Emerging morphologies in round bacterial colonies: comparing volumetric versus chemotactic expansion
Biomechanics and Modeling in Mechanobiology (2016)
-
PSF decomposition of nanoscopy images via Bayesian analysis unravels distinct molecular organization of the cell membrane
Scientific Reports (2014)