Abstract
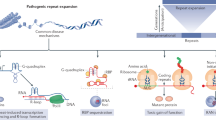
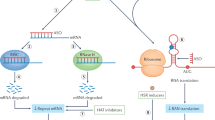
Expansions of repetitive DNA sequences cause numerous human neurological and neuromuscular diseases. Ongoing repeat expansions in patients can exacerbate disease progression and severity. As pathogenesis is connected to repeat length, a potential therapeutic avenue is to modulate disease by manipulating repeat expansion size — targeting DNA, the root-cause of symptoms. How repeat instability is mediated by DNA replication, repair, recombination, transcription and epigenetics may explain its contribution to pathogenesis and give insights into therapeutic strategies to block expansions or induce contractions.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Pearson, C. E., Nichol Edamura, K. & Cleary, J. D. Repeat instability: mechanisms of dynamic mutations. Nature Rev. Genet. 6, 729–742 (2005).
Swami, M. et al. Somatic expansion of the Huntington's disease CAG repeat in the brain is associated with an earlier age of disease onset. Hum. Mol. Genet. 18, 3039–3047 (2009).
Cleary, J. D. & Pearson, C. E. The contribution of cis-elements to disease-associated repeat instability: Clinical and experimental evidence. Cytogenet. Genome Res. 100, 25–55 (2003).
Gonitel, R. et al. DNA instability in postmitotic neurons. Proc. Natl Acad. Sci. USA 105, 3467–3472 (2008).
Entezam, A. et al. Regional FMRP deficits and large repeat expansions into the full mutation range in a new Fragile X premutation mouse model. Gene 395, 125–134 (2007).
Gomes-Pereira, M. et al. CTG trinucleotide repeat “big jumps”: large expansions, small mice. PLoS Genet. 3, e52 (2007).
Dion, V., Lin, Y., Hubert, L., Jr., Waterland, R. A. & Wilson, J. H. Dnmt1 deficiency promotes CAG repeat expansion in the mouse germline. Hum. Mol. Genet. 17, 1306–1317 (2008).
Entezam, A. & Usdin, K. ATM and ATR protect the genome against two different types of tandem repeat instability in Fragile X premutation mice. Nucleic Acids Res. 37, 6371–6377 (2009).
De Temmerman, N. et al. CTG repeat instability in a human embryonic stem cell line carrying the myotonic dystrophy type 1 mutation. Mol. Hum. Reprod. 14, 405–412 (2008).
Eiges, R. et al. Developmental study of fragile X syndrome using human embryonic stem cells derived from preimplantation genetically diagnosed embryos. Cell Stem Cell 1, 568–577 (2007).
Niclis, J. C. et al. Human embryonic stem cell models of Huntington disease. Reprod. Biomed. Online 19, 106–113 (2009).
Pearson, C. E. Slipping while sleeping? Trinucleotide repeat expansions in germ cells. Trends Mol. Med. 9, 490–495 (2003).
Voineagu, I., Freudenreich, C. H. & Mirkin, S. M. Checkpoint responses to unusual structures formed by DNA repeats. Mol. Carcinog. 48, 309–318 (2009).
Moe, S. E., Sorbo, J. G. & Holen, T. Huntingtin triplet-repeat locus is stable under long-term Fen1 knockdown in human cells. J. Neurosci. Methods 171, 233–238 (2008).
Lopez Castel, A., Tomkinson, A. E. & Pearson, C. E. CTG/CAG repeat instability is modulated by the levels of human DNA ligase I and its interaction with proliferating cell nuclear antigen: a distinction between replication and slipped-DNA repair. J. Biol. Chem. 284, 26631–26645 (2009).
Razidlo, D. F. & Lahue, R. S. Mrc1, Tof1 and Csm3 inhibit CAG•CTG repeat instability by at least two mechanisms. DNA Repair (Amst) 7, 633–640 (2008).
Shishkin, A. A. et al. Large-scale expansions of Friedreich's ataxia GAA repeats in yeast. Mol. Cell 35, 82–92 (2009).
Dhar, A. & Lahue, R. S. Rapid unwinding of triplet repeat hairpins by Srs2 helicase of Saccharomyces cerevisiae. Nucleic Acids Res. 36, 3366–3373 (2008).
Foiry, L. et al. Msh3 is a limiting factor in the formation of intergenerational CTG expansions in DM1 transgenic mice. Hum. Genet. 119, 520–526 (2006).
Owen, B. A. et al. (CAG)n-hairpin DNA binds to Msh2-Msh3 and changes properties of mismatch recognition. Nature Struct. Mol. Biol. 12, 663–670 (2005).
Tome, S. et al. MSH2 ATPase domain mutation affects CTG•CAG repeat instability in transgenic mice. PLoS Genet. 5, e1000482 (2009).
Slean, M. M., Panigrahi, G. B., Ranum, L. P. & Pearson, C. E. Mutagenic roles of DNA “repair” proteins in antibody diversity and disease-associated trinucleotide repeat instability. DNA Repair (Amst) 7, 1135–1154 (2008).
Hou, C., Chan, N. L., Gu, L. & Li, G. M. Incision-dependent and error-free repair of (CAG)n/(CTG)n hairpins in human cell extracts. Nature Struct. Mol. Biol. 16, 869–875 (2009).
Panigrahi, G. B., Lau, R., Montgomery, S. E., Leonard, M. R. & Pearson, C. E. Slipped (CTG)•(CAG) repeats can be correctly repaired, escape repair or undergo error-prone repair. Nature Struct. Mol. Biol. 12, 654–662 (2005).
Tian, L. et al. Mismatch recognition protein MutSβ does not hijack (CAG)n hairpin repair in vitro. J. Biol. Chem. 284, 20452–20456 (2009).
Liu, Y. et al. Coordination between polymerase β and FEN1 can modulate CAG repeat expansion. J. Biol. Chem. 284, 28352–28366 (2009).
Goula, A. V. et al. Stoichiometry of base excision repair proteins correlates with increased somatic CAG instability in striatum over cerebellum in Huntington's disease transgenic mice. PLoS Genet. 5, e1000749 (2009).
Jarem, D. A., Wilson, N. R. & Delaney, S. Structure-dependent DNA damage and repair in a trinucleotide repeat sequence. Biochemistry 48, 6655–6663 (2009).
Jung, J. & Bonini, N. CREB-binding protein modulates repeat instability in a Drosophila model for polyQ disease. Science 315, 1857–1859 (2007).
Lin, Y., Dion, V. & Wilson, J. H. Transcription promotes contraction of CAG repeat tracts in human cells. Nature Struct. Mol. Biol. 13, 179–180 (2006).
Lin, Y., Dent, S. Y., Wilson, J. H., Wells, R. D. & Napierala, M. R loops stimulate genetic instability of CTG•CAG repeats. Proc. Natl Acad. Sci. USA 107, 692–697 (2010).
Merienne, K. & Trottier, Y. SCA8 CAG/CTG expansions, a tale of two TOXICities: a unique or common case? PLoS Genet. 5, e1000593 (2009).
Ladd, P. D. et al. An antisense transcript spanning the CGG repeat region of FMR1 is upregulated in premutation carriers but silenced in full mutation individuals. Hum. Mol. Genet. 16, 3174–3187 (2007).
Kerrest, A. et al. SRS2 and SGS1 prevent chromosomal breaks and stabilize triplet repeats by restraining recombination. Nature Struct. Mol. Biol. 16, 159–167 (2009).
Libby, R. T. et al. CTCF cis-regulates trinucleotide repeat instability in an epigenetic manner: a novel basis for mutational hot spot determination. PLoS Genet. 4, e1000257 (2008).
Al-Mahdawi, S. et al. The Friedreich ataxia GAA repeat expansion mutation induces comparable epigenetic changes in human and transgenic mouse brain and heart tissues. Hum. Mol. Genet. 17, 735–746 (2008).
Castaldo, I. et al. DNA methylation in intron 1 of the frataxin gene is related to GAA repeat length and age of onset in Friedreich's ataxia patients. J. Med. Genet. 45, 808–812 (2008).
Edwards, S. F., Sirito, M., Krahe, R. & Sinden, R. R. A Z-DNA sequence reduces slipped-strand structure formation in the myotonic dystrophy type 2 (CCTG)•(CAGG) repeat. Proc. Natl Acad. Sci. USA 106, 3270–3275 (2009).
Kumari, D. & Usdin, K. Chromatin remodeling in the noncoding repeat expansion diseases. J. Biol. Chem. 284, 7413–7417 (2009).
Musova, Z. et al. Highly unstable sequence interruptions of the CTG repeat in the myotonic dystrophy gene. Am. J. Med. Genet. A 149A, 1365–1374 (2009).
Braida, C. et al. Variant CCG and GGC repeats within the CTG expansion dramatically modify mutational dynamics and likely contribute toward unusual symptoms in some myotonic dystrophy type 1 patients. Hum. Mol. Genet. 15 Jan 2010 (doi:10.1093/hmg/ddq015).
Sureshkumar, S. et al. A genetic defect caused by a triplet repeat expansion in Arabidopsis thaliana. Science 323, 1060–1063 (2009).
Lohi, H. et al. Expanded repeat in canine epilepsy. Science 307, 81 (2005).
Vinces, M. D., Legendre, M., Caldara, M., Hagihara, M. & Verstrepen, K. J. Unstable tandem repeats in promoters confer transcriptional evolvability. Science 324, 1213–1216 (2009).
Yang, Z., Lau, R., Marcadier, J. L., Chitayat, D. & Pearson, C. E. Replication inhibitors modulate instability of an expanded trinucleotide repeat at the myotonic dystrophy type 1 disease locus in human cells. Am. J. Hum. Genet. 73, 1092–1105 (2003).
Hashem, V. I. et al. Chemotherapeutic deletion of CTG repeats in lymphoblast cells from DM1 patients. Nucleic Acids Res. 32, 6334–6346 (2004).
Gomes-Pereira, M. & Monckton, D. G. Chemically induced increases and decreases in the rate of expansion of a CAG•CTG triplet repeat. Nucleic Acids Res. 32, 2865–2872 (2004).
Mittelman, D. et al. Zinc-finger directed double-strand breaks within CAG repeat tracts promote repeat instability in human cells. Proc. Natl Acad. Sci. USA 106, 9607–9612 (2009).
Mirkin, S. M. Expandable DNA repeats and human disease. Nature 447, 932–940 (2007).
Grabczyk, E., Mancuso, M. & Sammarco, M. C. A persistent RNA•DNA hybrid formed by transcription of the Friedreich ataxia triplet repeat in live bacteria, and by T7 RNAP in vitro. Nucleic Acids Res. 35, 5351–5359 (2007).
Acknowledgements
We apologize to colleagues whose work could not be cited owing to size limitations. Many citations are included in Supplementary information S1 and S2. Work in the Pearson laboratory is supported by the Muscular Dystrophy Association, USA, the Canadian Institutes of Health Research (CIHR) and the University of Rochester Paul Wellstone Muscular Dystrophy Cooperative Research Center, with support from the National Institutes of Health (U54NS48843), a CIHR fellowship (JDC) and the Hospital for Sick Children Research Training Centre (ALC).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Related links
Related links
DATABASES
OMIM
FURTHER INFORMATION
Christopher E. Pearson's homepage
Rights and permissions
About this article
Cite this article
López Castel, A., Cleary, J. & Pearson, C. Repeat instability as the basis for human diseases and as a potential target for therapy. Nat Rev Mol Cell Biol 11, 165–170 (2010). https://doi.org/10.1038/nrm2854
Issue Date:
DOI: https://doi.org/10.1038/nrm2854
This article is cited by
-
The mechanism of replication stalling and recovery within repetitive DNA
Nature Communications (2022)
-
Environmental exposures associated with elevated risk for autism spectrum disorder may augment the burden of deleterious de novo mutations among probands
Molecular Psychiatry (2022)
-
Krankheitsmodifizierende Therapieansätze bei der Huntington-Krankheit
Der Nervenarzt (2022)
-
Molecular mechanisms underlying nucleotide repeat expansion disorders
Nature Reviews Molecular Cell Biology (2021)
-
Conformational and migrational dynamics of slipped-strand DNA three-way junctions containing trinucleotide repeats
Nature Communications (2021)