Key Points
-
The ubiquitous influence of nitric oxide (NO) in cellular signalling is largely mediated by S-nitrosylation, the covalent attachment of NO to the thiol side chain of Cys residues. Proteins in most or all functional classes are substrates for S-nitrosylation, and a growing body of research shows that aberrant S-nitrosylation is implicated in a wide range of human pathologies.
-
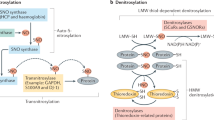
Recent discoveries indicate that the denitrosylation of proteins, which was once considered to be a spontaneous and unregulated event, is catalysed by enzymes in vivo. Denitrosylases might either directly mediate denitrosylation of proteins or govern the equilibrium between protein and low-molecular-weight nitrosothiols (collectively referred to as SNOs).
-
It has become increasingly clear that both S-nitrosylation and denitrosylation are precisely regulated in time and space. In particular, protein denitrosylation can be triggered by the stimulation of multiple classes of cell surface receptors, including members of the tumour necrosis factor family of receptors, G protein-coupled receptors and receptor Tyr kinases.
-
Several denitrosylases have recently been discovered, and two highly conserved enzyme systems in particular, the thioredoxin (Trx) system (which comprises Trx and Trx reductase (TrxR)) and the S-nitrosoglutathione reductase (GSNOR) system (which comprises glutathione (GSH) and GSNOR; GSNOR is also known as GSH-dependent formaldehyde dehydrogenase and class III alcohol dehydrogenase (ADH5) and is encoded by human gene ADH5) have been established to be physiologically relevant.
-
Trx proteins and GSNOR regulate the denitrosylation of multiple mammalian proteins and thereby modulate diverse cellular responses, including β-adrenergic receptor signalling, endocytosis, inflammation, angiogenesis and apoptotic cell death.
-
Denitrosylases are ubiquitously expressed in microbes and plants, in which they confer protection from nitrosative stress that is mediated by the host (that is, they serve as virulence factors) and exert profound effects on cellular immunity.
Abstract
S-Nitrosylation, the redox-based modification of Cys thiol side chains by nitric oxide, is a common mechanism in signal transduction. Dysregulated S-nitrosylation contributes to a range of human pathologies. New roles for protein denitrosylation in regulating S-nitrosylation are being revealed. Recently, several denitrosylases — the enzymes that mediate Cys denitrosylation — have been discovered, of which two enzyme systems in particular, the S-nitrosoglutathione reductase and thioredoxin systems, have been shown to be physiologically relevant. These highly conserved enzymes regulate signalling through multiple classes of receptors and influence diverse cellular responses. In addition, they protect from nitrosative stress in microorganisms, mammals and plants, thereby exerting profound effects on host–microbe interactions and innate immunity.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Hess, D. T., Matsumoto, A., Kim, S. O., Marshall, H. E. & Stamler, J. S. Protein S-nitrosylation: purview and parameters. Nature Rev. Mol. Cell Biol. 6, 150–166 (2005).
Jaffrey, S. R., Erdjument-Bromage, H., Ferris, C. D., Tempst, P. & Snyder, S. H. Protein S-nitrosylation: a physiological signal for neuronal nitric oxide. Nature Cell Biol. 3, 193–197 (2001).
Lane, P., Hao, G. & Gross, S. S. S-Nitrosylation is emerging as a specific and fundamental posttranslational protein modification: head-to-head comparison with O-phosphorylation. Sci. STKE 2001, RE1 (2001).
Durham, W. J. et al. RyR1 S-nitrosylation underlies environmental heat stroke and sudden death in Y522S RyR1 knockin mice. Cell 133, 53–65 (2008).
Bellinger, A. M. et al. Hypernitrosylated ryanodine receptor calcium release channels are leaky in dystrophic muscle. Nature Med. 15, 325–330 (2009).
Foster, M. W., McMahon, T. J. & Stamler, J. S. S-Nitrosylation in health and disease. Trends Mol. Med. 9, 160–168 (2003).
Hare, J. M. & Stamler, J. S. NO/redox disequilibrium in the failing heart and cardiovascular system. J. Clin. Invest. 115, 509–517 (2005).
Lim, K. H., Ancrile, B. B., Kashatus, D. F. & Counter, C. M. Tumour maintenance is mediated by eNOS. Nature 452, 646–649 (2008).
Uehara, T. et al. S-Nitrosylated protein-disulphide isomerase links protein misfolding to neurodegeneration. Nature 441, 513–517 (2006).
Guikema, B., Lu, Q. & Jourd'heuil, D. Chemical considerations and biological selectivity of protein nitrosation: implications for NO-mediated signal transduction. Antioxid. Redox Signal. 7, 593–606 (2005).
Foster, M. W., Liu, L., Zeng, M., Hess, D. T. & Stamler, J. S. A genetic analysis of nitrosative stress. Biochemistry 48, 792–799 (2009).
Singel, D. J. & Stamler, J. S. Chemical physiology of blood flow regulation by red blood cells: the role of nitric oxide and S-nitrosohemoglobin. Annu. Rev. Physiol. 67, 99–145 (2005).
Petersen, M. G., Dewilde, S. & Fago, A. Reactions of ferrous neuroglobin and cytoglobin with nitrite under anaerobic conditions. J. Inorg. Biochem. 102, 1777–1782 (2008).
Weichsel, A. et al. Heme-assisted S-nitrosation of a proximal thiolate in a nitric oxide transport protein. Proc. Natl Acad. Sci. USA 102, 594–599 (2005).
Inoue, K. et al. Nitrosothiol formation catalyzed by ceruloplasmin. Implication for cytoprotective mechanism in vivo. J. Biol. Chem. 274, 27069–27075 (1999).
Bosworth, C. A., Toledo, J. C. Jr, Zmijewski, J. W., Li, Q. & Lancaster, J. R. Jr. Dinitrosyliron complexes and the mechanism(s) of cellular protein nitrosothiol formation from nitric oxide. Proc. Natl Acad. Sci. USA 106, 4671–4676 (2009).
Gow, A. J. et al. Basal and stimulated protein S-nitrosylation in multiple cell types and tissues. J. Biol. Chem. 277, 9637–9640 (2002).
Fang, M. et al. Dexras1: a G protein specifically coupled to neuronal nitric oxide synthase via CAPON. Neuron 28, 183–193 (2000).
Rizzo, M. A. & Piston, D. W. Regulation of β cell glucokinase by S-nitrosylation and association with nitric oxide synthase. J. Cell Biol. 161, 243–248 (2003).
Kim, S. F., Huri, D. A. & Snyder, S. H. Inducible nitric oxide synthase binds, S-nitrosylates, and activates cyclooxygenase-2. Science 310, 1966–1970 (2005).
Iwakiri, Y. et al. Nitric oxide synthase generates nitric oxide locally to regulate compartmentalized protein S-nitrosylation and protein trafficking. Proc. Natl Acad. Sci. USA 103, 19777–19782 (2006).
Erwin, P. A., Mitchell, D. A., Sartoretto, J., Marletta, M. A. & Michel, T. Subcellular targeting and differential S-nitrosylation of endothelial nitric-oxide synthase. J. Biol. Chem. 281, 151–157 (2006).
Ozawa, K. et al. S-Nitrosylation of β-arrestin regulates β-adrenergic receptor trafficking. Mol. Cell 31, 395–405 (2008).
Hao, G., Derakhshan, B., Shi, L., Campagne, F. & Gross, S. S. SNOSID, a proteomic method for identification of cysteine S-nitrosylation sites in complex protein mixtures. Proc. Natl Acad. Sci. USA 103, 1012–1017 (2006).
Greco, T. M. et al. Identification of S-nitrosylation motifs by site-specific mapping of the S-nitrosocysteine proteome in human vascular smooth muscle cells. Proc. Natl Acad. Sci. USA 103, 7420–7425 (2006).
Mannick, J. B. et al. Fas-induced caspase denitrosylation. Science 284, 651–654 (1999). First demonstration of physiological protein denitrosylation.
Kim, J. E. & Tannenbaum, S. R. S-Nitrosation regulates the activation of endogenous procaspase-9 in HT-29 human colon carcinoma cells. J. Biol. Chem. 279, 9758–9764 (2004).
Erwin, P. A., Lin, A. J., Golan, D. E. & Michel, T. Receptor-regulated dynamic S-nitrosylation of endothelial nitric-oxide synthase in vascular endothelial cells. J. Biol. Chem. 280, 19888–19894 (2005). Demonstrates that denitrosylation is required for eNOS activity.
Reynaert, N. L. et al. Nitric oxide represses inhibitory κB kinase through S-nitrosylation. Proc. Natl Acad. Sci. USA 101, 8945–8950 (2004).
Forrester, M. T. et al. Proteomic analysis of S-nitrosylation and denitrosylation by resin-assisted capture. Nature Biotech. 27, 557–555 (2009). Identifies and characterizes protein denitrosylation on a global scale (in hundreds of proteins).
Stamler, J. S. & Toone, E. J. The decomposition of thionitrites. Curr. Opin. Chem. Biol. 6, 779–785 (2002).
Hogg, N. The biochemistry and physiology of S-nitrosothiols. Annu. Rev. Pharmacol. Toxicol. 42, 585–600 (2002).
Paige, J. S., Xu, G., Stancevic, B. & Jaffrey, S. R. Nitrosothiol reactivity profiling identifies S-nitrosylated proteins with unexpected stability. Chem. Biol. 15, 1307–1316 (2008). Proteomic analysis of glutathione-dependent denitrosylation.
Huang, B., Chen, S. C. & Wang, D. L. Shear flow increases S-nitrosylation of proteins in endothelial cells. Cardiovasc. Res. 83, 536–546 (2009).
Hoffmann, J., Haendeler, J., Zeiher, A. M. & Dimmeler, S. TNFα and oxLDL reduce protein S-nitrosylation in endothelial cells. J. Biol. Chem. 276, 41383–41387 (2001).
Chvanov, M., Gerasimenko, O. V., Petersen, O. H. & Tepikin, A. V. Calcium-dependent release of NO from intracellular S-nitrosothiols. EMBO J. 25, 3024–3032 (2006). The generation of NO by the GPCR agonist acetylcholine is mediated, in significant part, by protein denitrosylation.
Arnelle, D. R. & Stamler, J. S. NO+, NO˙, and NO− donation by S-nitrosothiols: implications for regulation of physiological functions by S-nitrosylation and acceleration of disulfide formation. Arch. Biochem. Biophys. 318, 279–285 (1995).
Barone, E. et al. Characterization of the S-denitrosylating activity of bilirubin. J. Cell. Mol. Med. 28 Jan 2009 (doi:10.1111/j.1582-4934.2008.00680.x).
Eu, J. P., Sun, J., Xu, L., Stamler, J. S. & Meissner, G. The skeletal muscle calcium release channel: coupled O2 sensor and NO signaling functions. Cell 102, 499–509 (2000).
Espey, M. G., Miranda, K. M., Thomas, D. D. & Wink, D. A. Distinction between nitrosating mechanisms within human cells and aqueous solution. J. Biol. Chem. 276, 30085–30091 (2001).
Foster, M. W. & Stamler, J. S. New insights into protein S-nitrosylation. Mitochondria as a model system. J. Biol. Chem. 279, 25891–25897 (2004).
Forrester, M. T., Foster, M. W. & Stamler, J. S. Assessment and application of the biotin switch technique for examining protein S-nitrosylation under conditions of pharmacologically induced oxidative stress. J. Biol. Chem. 282, 13977–13983 (2007).
Janssen-Heininger, Y. M. et al. Redox-based regulation of signal transduction: principles, pitfalls, and promises. Free Radic. Biol. Med. 45, 1–17 (2008).
Hewinson, J., Moore, S. F., Glover, C., Watts, A. G. & MacKenzie, A. B. A key role for redox signaling in rapid P2X7 receptor-induced IL-1β processing in human monocytes. J. Immunol. 180, 8410–8420 (2008).
Vogt, R. N., Steenkamp, D. J., Zheng, R. & Blanchard, J. S. The metabolism of nitrosothiols in the mycobacteria: identification and characterization of S-nitrosomycothiol reductase. Biochem. J. 374, 657–666 (2003).
Steffen, M. et al. Metabolism of S-nitrosoglutathione in intact mitochondria. Biochem. J. 356, 395–402 (2001).
Gordge, M. P., Addis, P., Noronha-Dutra, A. A. & Hothersall, J. S. Cell-mediated biotransformation of S-nitrosoglutathione. Biochem. Pharmacol. 55, 657–665 (1998).
Hausladen, A., Privalle, C. T., Keng, T., DeAngelo, J. & Stamler, J. S. Nitrosative stress: activation of the transcription factor OxyR. Cell 86, 719–729 (1996). The first report of cellular SNO lyase activity.
Hausladen, A., Gow, A. J. & Stamler, J. S. Nitrosative stress: metabolic pathway involving the flavohemoglobin. Proc. Natl Acad. Sci. USA 95, 14100–14105 (1998).
Nikitovic, D. & Holmgren, A. S-Nitrosoglutathione is cleaved by the thioredoxin system with liberation of glutathione and redox regulating nitric oxide. J. Biol. Chem. 271, 19180–19185 (1996). The first report of denitrosylation mediated by Trx/TrxR.
Trujillo, M., Alvarez, M. N., Peluffo, G., Freeman, B. A. & Radi, R. Xanthine oxidase-mediated decomposition of S-nitrosothiols. J. Biol. Chem. 273, 7828–7834 (1998).
Sliskovic, I., Raturi, A. & Mutus, B. Characterization of the S-denitrosation activity of protein disulfide isomerase. J. Biol. Chem. 280, 8733–8741 (2005).
Gaston, B. et al. Endogenous nitrogen oxides and bronchodilator S-nitrosothiols in human airways. Proc. Natl Acad. Sci. USA 90, 10957–10961 (1993).
Staab, C. A., Hellgren, M. & Hoog, J. O. Dual functions of alcohol dehydrogenase 3: implications with focus on formaldehyde dehydrogenase and S-nitrosoglutathione reductase activities. Cell. Mol. Life Sci. 65, 3950–3960 (2008).
Liu, L. et al. A metabolic enzyme for S-nitrosothiol conserved from bacteria to humans. Nature 410, 490–494 (2001).
Liu, L. et al. Essential roles of S-nitrosothiols in vascular homeostasis and endotoxic shock. Cell 116, 617–628 (2004). Together with reference 55, this study identifies the physiological roles of GSNOR.
Que, L. G. et al. Protection from experimental asthma by an endogenous bronchodilator. Science 308, 1618–1621 (2005).
Whalen, E. J. et al. Regulation of β-adrenergic receptor signaling by S-nitrosylation of G-protein-coupled receptor kinase 2. Cell 129, 511–522 (2007).
Wang, G., Moniri, N. H., Ozawa, K., Stamler, J. S. & Daaka, Y. Nitric oxide regulates endocytosis by S-nitrosylation of dynamin. Proc. Natl Acad. Sci. USA 103, 1295–1300 (2006).
Lima, B. et al. Endogenous S-nitrosothiols protect against myocardial injury. Proc. Natl Acad. Sci. USA 106, 6297–6302 (2009). The role of the GSNOR system in angiogenesis and myocardial protection.
Palmer, L. A. et al. S-Nitrosothiols signal hypoxia-mimetic vascular pathology. J. Clin. Invest. 117, 2592–2601 (2007).
Lipton, A. J. et al. S-Nitrosothiols signal the ventilatory response to hypoxia. Nature 413, 171–174 (2001).
Dimmeler, S., Haendeler, J., Nehls, M. & Zeiher, A. M. Suppression of apoptosis by nitric oxide via inhibition of interleukin-1β-converting enzyme (ICE)-like and cysteine protease protein (CPP)-32-like proteases. J. Exp. Med. 185, 601–607 (1997).
Kim, Y. M., Talanian, R. V. & Billiar, T. R. Nitric oxide inhibits apoptosis by preventing increases in caspase-3-like activity via two distinct mechanisms. J. Biol. Chem. 272, 31138–31148 (1997).
Rossig, L. et al. Nitric oxide inhibits caspase-3 by S-nitrosation in vivo. J. Biol. Chem. 274, 6823–6826 (1999). Early demonstration of physiological S -nitrosylation of caspase 3.
Mannick, J. B. et al. S-Nitrosylation of mitochondrial caspases. J. Cell Biol. 154, 1111–1116 (2001). Documents compartmentalization of S-nitrosylated caspases.
Lillig, C. H. & Holmgren, A. Thioredoxin and related molecules-from biology to health and disease. Antioxid. Redox Signal. 9, 25–47 (2007).
Nikitovic, D., Holmgren, A. & Spyrou, G. Inhibition of AP-1 DNA binding by nitric oxide involving conserved cysteine residues in Jun and Fos. Biochem. Biophys. Res. Commun. 242, 109–112 (1998).
Kahlos, K., Zhang, J., Block, E. R. & Patel, J. M. Thioredoxin restores nitric oxide-induced inhibition of protein kinase C activity in lung endothelial cells. Mol. Cell. Biochem. 254, 47–54 (2003).
Zhang, J., Li, Y. D., Patel, J. M. & Block, E. R. Thioredoxin overexpression prevents NO-induced reduction of NO synthase activity in lung endothelial cells. Am. J. Physiol. 275, L288–L293 (1998).
Ravi, K., Brennan, L. A., Levic, S., Ross, P. A. & Black, S. M. S-Nitrosylation of endothelial nitric oxide synthase is associated with monomerization and decreased enzyme activity. Proc. Natl Acad. Sci. USA 101, 2619–2624 (2004).
Stoyanovsky, D. A. et al. Thioredoxin and lipoic acid catalyze the denitrosation of low molecular weight and protein S-Nitrosothiols. J. Am. Chem. Soc. 127, 15815–15823 (2005).
Sengupta, R. et al. Thioredoxin catalyzes the denitrosation of low-molecular mass and protein S-nitrosothiols. Biochemistry 46, 8472–8483 (2007).
Benhar, M., Forrester, M. T., Hess, D. T. & Stamler, J. S. Regulated protein denitrosylation by cytosolic and mitochondrial thioredoxins. Science 320, 1050–1054 (2008). Identification of Trx and TrxR proteins as physiological protein denitrosylases.
Kang, S. W., Rhee, S. G., Chang, T. S., Jeong, W. & Choi, M. H. 2-Cys peroxiredoxin function in intracellular signal transduction: therapeutic implications. Trends Mol. Med. 11, 571–578 (2005).
Spyrou, G., Enmark, E., Miranda-Vizuete, A. & Gustafsson, J. Cloning and expression of a novel mammalian thioredoxin. J. Biol. Chem. 272, 2936–2941 (1997).
Chanvorachote, P. et al. Nitric oxide negatively regulates Fas CD95-induced apoptosis through inhibition of ubiquitin-proteasome-mediated degradation of FLICE inhibitory protein. J. Biol. Chem. 280, 42044–42050 (2005).
Azad, N. et al. S-Nitrosylation of Bcl-2 inhibits its ubiquitin-proteasomal degradation. A novel antiapoptotic mechanism that suppresses apoptosis. J. Biol. Chem. 281, 34124–34134 (2006).
Mitchell, D. A., Morton, S. U., Fernhoff, N. B. & Marletta, M. A. Thioredoxin is required for S-nitrosation of procaspase-3 and the inhibition of apoptosis in Jurkat cells. Proc. Natl Acad. Sci. USA 104, 11609–11614 (2007).
Chanvorachote, P. et al. Nitric oxide regulates cell sensitivity to cisplatin-induced apoptosis through S-nitrosylation and inhibition of Bcl-2 ubiquitination. Cancer Res. 66, 6353–6360 (2006).
Haendeler, J. et al. Redox regulatory and anti-apoptotic functions of thioredoxin depend on S-nitrosylation at cysteine 69. Nature Cell Biol. 4, 743–749 (2002).
Papadia, S. et al. Synaptic NMDA receptor activity boosts intrinsic antioxidant defenses. Nature Neurosci. 11, 476–487 (2008).
Schulze, P. C. et al. Nitric oxide-dependent suppression of thioredoxin-interacting protein expression enhances thioredoxin activity. Arterioscler. Thromb. Vasc. Biol. 26, 2666–2672 (2006).
Andoh, T., Chiueh, C. C. & Chock, P. B. Cyclic GMP-dependent protein kinase regulates the expression of thioredoxin and thioredoxin peroxidase-1 during hormesis in response to oxidative stress-induced apoptosis. J. Biol. Chem. 278, 885–890 (2003).
Hromatka, B. S., Noble, S. M. & Johnson, A. D. Transcriptional response of Candida albicans to nitric oxide and the role of the YHB1 gene in nitrosative stress and virulence. Mol. Biol. Cell 16, 4814–4826 (2005).
Sumbayev, V. V. S-Nitrosylation of thioredoxin mediates activation of apoptosis signal-regulating kinase 1. Arch. Biochem. Biophys. 415, 133–136 (2003).
Tao, L. et al. Cardioprotective effects of thioredoxin in myocardial ischemia and reperfusion: role of S-nitrosation. Proc. Natl Acad. Sci. USA 101, 11471–11476 (2004).
Yasinska, I. M., Kozhukhar, A. V. & Sumbayev, V. V. S-Nitrosation of thioredoxin in the nitrogen monoxide/superoxide system activates apoptosis signal-regulating kinase 1. Arch. Biochem. Biophys. 428, 198–203 (2004).
Mitchell, D. A. & Marletta, M. A. Thioredoxin catalyzes the S-nitrosation of the caspase-3 active site cysteine. Nature Chem. Biol. 1, 154–158 (2005).
Weichsel, A., Brailey, J. L. & Montfort, W. R. Buried S-nitrosocysteine revealed in crystal structures of human thioredoxin. Biochemistry 46, 1219–1227 (2007).
Hashemy, S. I. & Holmgren, A. Regulation of the catalytic activity and structure of human thioredoxin 1 via oxidation and S-nitrosylation of cysteine residues. J. Biol. Chem. 283, 21890–21898 (2008).
Stewart, E. J., Aslund, F. & Beckwith, J. Disulfide bond formation in the Escherichia coli cytoplasm: an in vivo role reversal for the thioredoxins. EMBO. J. 17, 5543–5550 (1998).
Bhandari, V. et al. Essential role of nitric oxide in VEGF-induced, asthma-like angiogenic, inflammatory, mucus, and physiologic responses in the lung. Proc. Natl Acad. Sci. USA 103, 11021–11026 (2006).
Lopez-Sanchez, L. M. et al. Alteration of S-nitrosothiol homeostasis and targets for protein S-nitrosation in human hepatocytes. Proteomics 8, 4709–4720 (2008).
Kidd, S. P., Jiang, D., Jennings, M. P. & McEwan, A. G. Glutathione-dependent alcohol dehydrogenase AdhC is required for defense against nitrosative stress in Haemophilus influenzae. Infect. Immun. 75, 4506–4513 (2007).
Stroeher, U. H. et al. A pneumococcal MerR-like regulator and S-nitrosoglutathione reductase are required for systemic virulence. J. Infect. Dis. 196, 1820–1826 (2007).
de Jesus-Berrios, M. et al. Enzymes that counteract nitrosative stress promote fungal virulence. Curr. Biol. 13, 1963–1968 (2003). References 95–97 describe important roles for GSNOR in pathogen survival and virulence.
Bang, I. S. et al. Maintenance of nitric oxide and redox homeostasis by the Salmonella flavohemoglobin Hmp. J. Biol. Chem. 281, 28039–28047 (2006).
Comtois, S. L., Gidley, M. D. & Kelly, D. J. Role of the thioredoxin system and the thiol-peroxidases Tpx and Bcp in mediating resistance to oxidative and nitrosative stress in Helicobacter pylori. Microbiology 149, 121–129 (2003).
Potter, A. J. et al. Thioredoxin reductase is essential for protection of Neisseria gonorrhoeae against killing by nitric oxide and for bacterial growth during interaction with cervical epithelial cells. J. Infect. Dis. 199, 227–235 (2009).
Eu, J. P., Liu, L., Zeng, M. & Stamler, J. S. An apoptotic model for nitrosative stress. Biochemistry 39, 1040–1047 (2000).
Hara, M. R. et al. S-Nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nature Cell Biol. 7, 665–674 (2005).
Benhar, M. & Stamler, J. S. A central role for S-nitrosylation in apoptosis. Nature Cell Biol. 7, 645–646 (2005).
Wang, Y. et al. S-Nitrosylation: an emerging redox-based post-translational modification in plants. J. Exp. Bot. 57, 1777–1784 (2006).
Feechan, A. et al. A central role for S-nitrosothiols in plant disease resistance. Proc. Natl Acad. Sci. USA 102, 8054–8059 (2005).
Tada, Y. et al. Plant immunity requires conformational charges of NPR1 via S-nitrosylation and thioredoxins. Science 321, 952–956 (2008).
Wang, Y. Q. et al. S-Nitrosylation of AtSABP3 antagonizes the expression of plant immunity. J. Biol. Chem. 284, 2131–2137 (2009). References 105–107 demonstrate important functions of GSNOR in plant immunity.
Marshall, H. E. & Stamler, J. S. Inhibition of NF-κB by S-nitrosylation. Biochemistry 40, 1688–1693 (2001).
Into, T. et al. Regulation of MyD88-dependent signaling events by S nitrosylation retards toll-like receptor signal transduction and initiation of acute-phase immune responses. Mol. Cell. Biol. 28, 1338–1347 (2008).
Ferret, P. J., Soum, E., Negre, O., Wollman, E. E. & Fradelizi, D. Protective effect of thioredoxin upon NO-mediated cell injury in THP1 monocytic human cells. Biochem. J. 346, 759–765 (2000).
Arai, R. J. et al. Thioredoxin-1 promotes survival in cells exposed to S-nitrosoglutathione: correlation with reduction of intracellular levels of nitrosothiols and up-regulation of the ERK1/2 MAP kinases. Toxicol. Appl. Pharmacol. 233, 227–237 (2008).
Zai, A., Rudd, M. A., Scribner, A. W. & Loscalzo, J. Cell-surface protein disulfide isomerase catalyzes transnitrosation and regulates intracellular transfer of nitric oxide. J. Clin. Invest. 103, 393–399 (1999).
Jourd'heuil, D., Laroux, F. S., Miles, A. M., Wink, D. A. & Grisham, M. B. Effect of superoxide dismutase on the stability of S-nitrosothiols. Arch. Biochem. Biophys. 361, 323–330 (1999).
Johnson, M. A., Macdonald, T. L., Mannick, J. B., Conaway, M. R. & Gaston, B. Accelerated S-nitrosothiol breakdown by amyotrophic lateral sclerosis mutant copper, zinc-superoxide dismutase. J. Biol. Chem. 276, 39872–39878 (2001).
Schonhoff, C. M. et al. S-Nitrosothiol depletion in amyotrophic lateral sclerosis. Proc. Natl Acad. Sci. USA 103, 2404–2409 (2006).
Okado-Matsumoto, A. & Fridovich, I. Putative denitrosylase activity of Cu, Zn-superoxide dismutase. Free Radic. Biol. Med. 43, 830–836 (2007).
Romeo, A. A., Capobianco, J. A. & English, A. M. Superoxide dismutase targets NO from GSNO to Cysβ93 of oxyhemoglobin in concentrated but not dilute solutions of the protein. J. Am. Chem. Soc. 125, 14370–14378 (2003).
Hou, Y., Guo, Z., Li, J. & Wang, P. G. Seleno compounds and glutathione peroxidase catalyzed decomposition of S-nitrosothiols. Biochem. Biophys. Res. Commun. 228, 88–93 (1996).
Freedman, J. E., Frei, B., Welch, G. N. & Loscalzo, J. Glutathione peroxidase potentiates the inhibition of platelet function by S-nitrosothiols. J. Clin. Invest. 96, 394–400 (1995).
Bateman, R. L., Rauh, D., Tavshanjian, B. & Shokat, K. M. Human carbonyl reductase 1 is an S-nitrosoglutathione reductase. J. Biol. Chem. 283, 35756–35762 (2008). The most recent characterization of a novel Cys denitrosylase.
Pawloski, J. R., Hess, D. T. & Stamler, J. S. Export by red blood cells of nitric oxide bioactivity. Nature 409, 622–626 (2001).
Ishima, Y. et al. S-Nitrosylated human serum albumin-mediated cytoprotective activity is enhanced by fatty acid binding. J. Biol. Chem. 283, 34966–34975 (2008).
Jourd'heuil, D., Mai, C. T., Laroux, F. S., Wink, D. A. & Grisham, M. B. The reaction of S-nitrosoglutathione with superoxide. Biochem. Biophys. Res. Commun. 246, 525–530 (1998).
Cheng, F. et al. Nitric oxide-dependent processing of heparan sulfate in recycling S-nitrosylated glypican-1 takes place in caveolin-1-containing endosomes. J. Biol. Chem. 277, 44431–44439 (2002).
Heinrich, R., Neel, B. G. & Rapoport, T. A. Mathematical models of protein kinase signal transduction. Mol. Cell 9, 957–970 (2002).
Pawloski, J. R., Hess, D. T. & Stamler, J. S. Impaired vasodilation by red blood cells in sickle cell disease. Proc. Natl Acad. Sci. USA 102, 2531–2536 (2005).
McMahon, T. J. et al. A nitric oxide processing defect of red blood cells created by hypoxia: deficiency of S-nitrosohemoglobin in pulmonary hypertension. Proc. Natl Acad. Sci. USA 102, 14801–14806 (2005).
Wu, H. et al. Genetic variation in S-nitrosoglutathione reductase (GSNOR) and childhood asthma. J. Allergy Clin. Immunol. 120, 322–328 (2007).
Davisson, R. L., Bates, J. N., Johnson, A. K. & Lewis, S. J. Use-dependent loss of acetylcholine- and bradykinin-mediated vasodilation after nitric oxide synthase inhibition. Evidence for preformed stores of nitric oxide-containing factors in vascular endothelial cells. Hypertension 28, 354–360 (1996).
Rhee, K. Y., Erdjument-Bromage, H., Tempst, P. & Nathan, C. F. S-Nitroso proteome of Mycobacterium tuberculosis: enzymes of intermediary metabolism and antioxidant defense. Proc. Natl Acad. Sci. USA 102, 467–472 (2005).
Fang, F. C. Antimicrobial reactive oxygen and nitrogen species: concepts and controversies. Nature Rev. Microbiol. 2, 820–832 (2004).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors are inventors on patents that concern protein denitrosylation, and J. Stamler is involved in the development of nitric oxide-based technologies.
Related links
Related links
DATABASES
Entrez Gene
TAIR
FURTHER INFORMATION
Glossary
- G protein-coupled receptor
-
A cell surface receptor (such as the β-adrenergic and the cholinergic receptors) that possesses seven transmembrane domains and is coupled to G proteins. Typically, the activation of a G protein-coupled receptor produces a second messenger that initiates a signal transduction cascade.
- NO synthase
-
(NOS). Mammals have three nitric oxide (NO) synthases that generate NO from Arg NOS1 (or neuronal NOS), NOS2 (or inducible NOS) and NOS3 (or endothelial NOS) one or more of which reside or can be induced in most or all cell types. NOS orthologues are distributed broadly across phylogeny.
- Metalloprotein
-
A protein that contains a metal ion or ions (including Fe2+, Cu2+ or Zn2+) as a prosthetic group that is coordinated by amino acid side chains.
- Nitrosative stress
-
The dysregulated production and/or metabolism of reactive nitrogen species, which generate nitrosative chemistries that can result in disrupted cellular signalling, injury and death. Oxidative stress is brought about by reactive oxygen species.
- S-Nitrosoglutathione
-
The main non-protein S-nitrosothiol (SNO) in cells, which can be present in micromolar concentrations, and that is in equilibrium with protein SNOs.
- Glutathione
-
A tripeptide composed of Glu, Cys and Gly that is the principal, small-molecular-weight, thiol-containing molecule in the cell.
- Nucleophilic
-
Having an affinity for positive charge. Nucleophilic molecules are electron rich and tend to attack electron-poor molecules or behave as electron donors.
- Reactive oxygen species
-
Reduced derivatives of molecular oxygen that include, in particular, the superoxide radical (O2−) and hydrogen peroxide (H2O2), which can have substantial reactivity towards biological macromolecules and towards other reactive small molecules.
- Lyase
-
Typically, an enzyme that breaks a bond without hydrolysis or oxidation.
- NADH
-
The reduced form of nicotinamide adenine dinucleotide. This coenzyme serves as an electron donor for various biochemical reactions.
- Caspase
-
A family of Cys proteases, divided into initiator and effector caspases, that might require proteolytic cleavage to liberate subunits that reconstitute an active caspase heterodimer. All caspases contain a Cys residue at the active site and cleave substrates carboxy-terminal to an Asp residue.
- Oxidoreductase
-
An enzyme that catalyses oxidation–reduction reactions. These entail the transfer of electrons from a substrate that becomes oxidized (electron donor) to a substrate that becomes reduced (electron acceptor).
- Death receptor
-
One of a family of cell surface receptors that mediate cell death upon ligand-induced trimerization. The best-studied members include tumour necrosis factor receptor 1 (TNFR1) and FAS (or CD95), which binds the FAS ligand.
- Virulence
-
The damage caused to the host by a parasite or pathogen, which is measured as a decrease in host fitness.
Rights and permissions
About this article
Cite this article
Benhar, M., Forrester, M. & Stamler, J. Protein denitrosylation: enzymatic mechanisms and cellular functions. Nat Rev Mol Cell Biol 10, 721–732 (2009). https://doi.org/10.1038/nrm2764
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrm2764
This article is cited by
-
Nitric oxide-driven modifications of lipoic arm inhibit α-ketoacid dehydrogenases
Nature Chemical Biology (2023)
-
S-nitrosylation of c-Jun N-terminal kinase mediates pressure overload-induced cardiac dysfunction and fibrosis
Acta Pharmacologica Sinica (2022)
-
Insights into the post-translational modification and its emerging role in shaping the tumor microenvironment
Signal Transduction and Targeted Therapy (2021)
-
Shank3 mutation in a mouse model of autism leads to changes in the S-nitroso-proteome and affects key proteins involved in vesicle release and synaptic function
Molecular Psychiatry (2020)
-
A Novel Mechanism for Nitrosative Stress Tolerance Dependent on GTP Cyclohydrolase II Activity Involved in Riboflavin Synthesis of Yeast
Scientific Reports (2020)