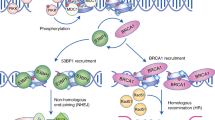
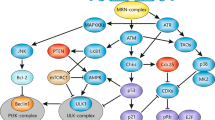
Key Points
-
The repair of DNA lesions that occur endogenously or in response to diverse genotoxic stresses is indispensable for maintaining genome integrity.
-
The types of DNA lesion and the checkpoint pathways that are activated in response to DNA damage influence the DNA-repair pathways according to the cell-cycle phase.
-
Failure to coordinate DNA repair with cell-cycle progression can cause genome instability, cell death and cancer.
-
Phosphorylation events that are mediated by cyclin-dependent kinases and checkpoints regulate DNA repair according to the cell-cycle stage.
-
Certain DNA-repair pathways are attenuated in non-dividing cells that probably possess dedicated mechanisms to repair endogenous lesions.
-
SUMO and ubiquitin modifications are crucial in the regulation of the stability and activity of key components of DNA repair and checkpoint machineries, thereby regulating important cell-cycle events.
Abstract
The repair of DNA lesions that occur endogenously or in response to diverse genotoxic stresses is indispensable for genome integrity. DNA lesions activate checkpoint pathways that regulate specific DNA-repair mechanisms in the different phases of the cell cycle. Checkpoint-arrested cells resume cell-cycle progression once damage has been repaired, whereas cells with unrepairable DNA lesions undergo permanent cell-cycle arrest or apoptosis. Recent studies have provided insights into the mechanisms that contribute to DNA repair in specific cell-cycle phases and have highlighted the mechanisms that ensure cell-cycle progression or arrest in normal and cancerous cells.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Sancar, A., Lindsey-Boltz, L. A., Unsal-Kacmaz, K. & Linn, S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 73, 39–85 (2004).
Vermeulen, K., Van Bockstaele, D. R. & Berneman, Z. N. The cell cycle: a review of regulation, deregulation and therapeutic targets in cancer. Cell Prolif. 36, 131–149 (2003).
Aguilera, A. The connection between transcription and genomic instability. EMBO J. 21, 195–201 (2002).
Branzei, D. & Foiani, M. The DNA damage response during DNA replication. Curr. Opin. Cell Biol. 17, 568–575 (2005).
Strom, L. & Sjogren, C. Chromosome segregation and double-strand break repair — a complex connection. Curr. Opin. Cell Biol. 19, 344–349 (2007).
Wang, J. C. Cellular roles of DNA topoisomerases: a molecular perspective. Nature Rev. Mol. Cell Biol. 3, 430–440 (2002).
Branzei, D. & Foiani, M. The Rad53 signal transduction pathway: replication fork stabilization, DNA repair, and adaptation. Exp. Cell Res. 312, 2654–2659 (2006).
Bartek, J. & Lukas, J. DNA damage checkpoints: from initiation to recovery or adaptation. Curr. Opin. Cell Biol. 19, 238–245 (2007).
Neecke, H., Lucchini, G. & Longhese, M. P. Cell cycle progression in the presence of irreparable DNA damage is controlled by a Mec1- and Rad53-dependent checkpoint in budding yeast. EMBO J. 18, 4485–4497 (1999).
Giannattasio, M., Lazzaro, F., Longhese, M. P., Plevani, P. & Muzi-Falconi, M. Physical and functional interactions between nucleotide excision repair and DNA damage checkpoint. EMBO J. 23, 429–438 (2004).
Pellicioli, A., Lee, S. E., Lucca, C., Foiani, M. & Haber, J. E. Regulation of Saccharomyces Rad53 checkpoint kinase during adaptation from DNA damage-induced G2/M arrest. Mol. Cell 7, 293–300 (2001).
Ira, G. et al. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature 431, 1011–1017 (2004). Presents the first evidence that CDK1 activity is required for DSB end resection and so influences the choice of the DSBR pathway according to the cell-cycle phase.
Jazayeri, A. et al. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nature Cell Biol. 8, 37–45 (2006). Shows a dependency of ATR activation on ATM, MRN and CDK activity, and demonstrates the link between ATR activation and HR repair.
Cotta-Ramusino, C. et al. Exo1 processes stalled replication forks and counteracts fork reversal in checkpoint-defective cells. Mol. Cell 17, 153–159 (2005).
Elledge, S. J. Cell cycle checkpoints: preventing an identity crisis. Science 274, 1664–1672 (1996).
Falck, J., Coates, J. & Jackson, S. P. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature 434, 605–611 (2005).
You, Z., Chahwan, C., Bailis, J., Hunter, T. & Russell, P. ATM activation and its recruitment to damaged DNA require binding to the C terminus of Nbs1. Mol. Cell Biol. 25, 5363–5379 (2005).
Aylon, Y., Liefshitz, B. & Kupiec, M. The CDK regulates repair of double-strand breaks by homologous recombination during the cell cycle. EMBO J. 23, 4868–4875 (2004).
Matsuoka, S. et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 316, 1160–1166 (2007). Identifies a comprehensive catalogue of ATM and ATR targets and provides evidence that they are bona fide members of the DDR network.
Kouzarides, T. Chromatin modifications and their function. Cell 128, 693–705 (2007).
Karagiannis, T. C. & El-Osta, A. Chromatin modifications and DNA double-strand breaks: the current state of play. Leukemia 21, 195–200 (2007).
Groth, A., Rocha, W., Verreault, A. & Almouzni, G. Chromatin challenges during DNA replication and repair. Cell 128, 721–733 (2007).
Shibutani, S., Takeshita, M. & Grollman, A. P. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature 349, 431–434 (1991).
Soussi, T. & Beroud, C. Significance of TP53 mutations in human cancer: a critical analysis of mutations at CpG dinucleotides. Hum. Mutat. 21, 192–200 (2003).
Russo, M. T. et al. Accumulation of the oxidative base lesion 8-hydroxyguanine in DNA of tumor-prone mice defective in both the Myh and Ogg1 DNA glycosylases. Cancer Res. 64, 4411–4414 (2004).
Nojima, K. et al. Multiple repair pathways mediate tolerance to chemotherapeutic cross-linking agents in vertebrate cells. Cancer Res. 65, 11704–11711 (2005).
Sonoda, E., Hochegger, H., Saberi, A., Taniguchi, Y. & Takeda, S. Differential usage of non-homologous end-joining and homologous recombination in double strand break repair. DNA Repair 5, 1021–1029 (2006).
Paques, F. & Haber, J. E. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63, 349–404 (1999).
Krogh, B. O. & Symington, L. S. Recombination proteins in yeast. Annu. Rev. Genet. 38, 233–271 (2004).
Takata, M. et al. Homologous recombination and non-homologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J. 17, 5497–5508 (1998).
Lee, S. E., Mitchell, R. A., Cheng, A. & Hendrickson, E. A. Evidence for DNA-PK-dependent and -independent DNA double-strand break repair pathways in mammalian cells as a function of the cell cycle. Mol. Cell Biol. 17, 1425–1433 (1997).
Astrom, S. U., Okamura, S. M. & Rine, J. Yeast cell-type regulation of DNA repair. Nature 397, 310 (1999).
Lee, S. E., Paques, F., Sylvan, J. & Haber, J. E. Role of yeast SIR genes and mating type in directing DNA double-strand breaks to homologous and non-homologous repair paths. Curr. Biol. 9, 767–770 (1999).
Jiricny, J. The multifaceted mismatch-repair system. Nature Rev. Mol. Cell Biol. 7, 335–346 (2006).
Lettier, G. et al. The role of DNA double-strand breaks in spontaneous homologous recombination in S. cerevisiae. PLoS Genet. 2, e194 (2006).
Fabre, F., Chan, A., Heyer, W. D. & Gangloff, S. Alternate pathways involving Sgs1/Top3, Mus81/ Mms4, and Srs2 prevent formation of toxic recombination intermediates from single-stranded gaps created by DNA replication. Proc. Natl Acad. Sci. USA 99, 16887–16892 (2002).
Branzei, D. & Foiani, M. Interplay of replication checkpoints and repair proteins at stalled replication forks. DNA Repair 6, 994–1003 (2007).
Lehmann, A. R. et al. Translesion synthesis: Y-family polymerases and the polymerase switch. DNA Repair 6, 891–899 (2007).
Hoege, C., Pfander, B., Moldovan, G. L., Pyrowolakis, G. & Jentsch, S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419, 135–141 (2002). Presents the first evidence that PCNA is modified by ubiquitin and SUMO and demonstrates that PCNA polyubiquitylation is required for PRR.
Sung, P. & Klein, H. Mechanism of homologous recombination: mediators and helicases take on regulatory functions. Nature Rev. Mol. Cell Biol. 7, 739–750 (2006).
Nakada, D., Hirano, Y. & Sugimoto, K. Requirement of the Mre11 complex and exonuclease 1 for activation of the Mec1 signaling pathway. Mol. Cell Biol. 24, 10016–10025 (2004).
Llorente, B. & Symington, L. S. The Mre11 nuclease is not required for 5′ to 3′ resection at multiple HO-induced double-strand breaks. Mol. Cell Biol. 24, 9682–9694 (2004).
Limbo, O. et al. Ctp1 is a cell-cycle-regulated protein that functions with Mre11 complex to control double-strand break repair by homologous recombination. Mol. Cell 28, 134–146 (2007). Together with reference 44, this paper documents the ability of Ctp1 (or CtIP in mammals) to function together with the MRX (or MRN in mammals) complex in DSB resection and promote HR in the S and G2 phases of the cell cycle.
Sartori, A. A. et al. Human CtIP promotes DNA end resection. Nature 450, 509–514 (2007).
Aylon, Y. & Kupiec, M. New insights into the mechanism of homologous recombination in yeast. Mutat. Res. 566, 231–248 (2004).
Kim, J. S. et al. Independent and sequential recruitment of NHEJ and HR factors to DNA damage sites in mammalian cells. J. Cell Biol. 170, 341–347 (2005).
Hochegger, H. et al. PARP-1 protects homologous recombination from interference by Ku and Ligase IV in vertebrate cells. EMBO J. 25, 1305–1314 (2006).
Saberi, A. et al. RAD18 and poly(ADP-ribose) polymerase independently suppress the access of nonhomologous end joining to double-strand breaks and facilitate homologous recombination-mediated repair. Mol. Cell Biol. 27, 2562–2571 (2007).
Barber, L. J. & Boulton, S. J. BRCA1 ubiquitylation of CtIP: just the tIP of the iceberg? DNA Repair 5, 1499–1504 (2006).
Yu, X. & Baer, R. Nuclear localization and cell cycle-specific expression of CtIP, a protein that associates with the BRCA1 tumor suppressor. J. Biol. Chem. 275, 18541–18549 (2000).
Hirano, T. At the heart of the chromosome: SMC proteins in action. Nature Rev. Mol. Cell Biol. 7, 311–322 (2006).
Uhlmann, F. & Nasmyth, K. Cohesion between sister chromatids must be established during DNA replication. Curr. Biol. 8, 1095–1101 (1998).
Skibbens, R. V., Corson, L. B., Koshland, D. & Hieter, P. Ctf7p is essential for sister chromatid cohesion and links mitotic chromosome structure to the DNA replication machinery. Genes Dev. 13, 307–319 (1999).
Strom, L. et al. Postreplicative formation of cohesion is required for repair and induced by a single DNA break. Science 317, 242–245 (2007).
Unal, E., Heidinger-Pauli, J. M. & Koshland, D. DNA double-strand breaks trigger genome-wide sister-chromatid cohesion through Eco1 (Ctf7). Science 317, 245–248 (2007).
Sjogren, C. & Nasmyth, K. Sister chromatid cohesion is required for postreplicative double-strand break repair in Saccharomyces cerevisiae. Curr. Biol. 11, 991–995 (2001).
Strom, L., Lindroos, H. B., Shirahige, K. & Sjogren, C. Postreplicative recruitment of cohesin to double-strand breaks is required for DNA repair. Mol. Cell 16, 1003–1015 (2004).
Unal, E. et al. DNA damage response pathway uses histone modification to assemble a double-strand break-specific cohesin domain. Mol. Cell 16, 991–1002 (2004).
Bermejo, R. et al. Top1- and Top2-mediated topological transitions at replication forks ensure fork progression and stability and prevent DNA damage checkpoint activation. Genes Dev. 21, 1921–1936 (2007).
Franchitto, A., Oshima, J. & Pichierri, P. The G2-phase decatenation checkpoint is defective in Werner syndrome cells. Cancer Res. 63, 3289–3295 (2003).
Deming, P. B. et al. The human decatenation checkpoint. Proc. Natl Acad. Sci. USA 98, 12044–12049 (2001).
Deming, P. B., Flores, K. G., Downes, C. S., Paules, R. S. & Kaufmann, W. K. ATR enforces the topoisomerase II-dependent G2 checkpoint through inhibition of Plk1 kinase. J. Biol. Chem. 277, 36832–36838 (2002).
Nouspikel, T. DNA repair in differentiated cells: some new answers to old questions. Neuroscience 145, 1213–1221 (2007).
Wilson, D. M. 3rd & McNeill, D. R. Base excision repair and the central nervous system. Neuroscience 145, 1187–1200 (2007).
Fishel, M. L., Vasko, M. R. & Kelley, M. R. DNA repair in neurons: so if they don't divide what's to repair? Mutat. Res. 614, 24–36 (2007).
Lindahl, T., Karran, P. & Wood, R. D. DNA excision repair pathways. Curr. Opin. Genet. Dev. 7, 158–169 (1997).
Lisby, M., Barlow, J. H., Burgess, R. C. & Rothstein, R. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell 118, 699–713 (2004).
Yuan, S. S., Chang, H. L. & Lee, E. Y. Ionizing radiation-induced Rad51 nuclear focus formation is cell cycle-regulated and defective in both ATM−/− and c-Abl−/− cells. Mutat. Res. 525, 85–92 (2003).
Morrison, C. et al. The controlling role of ATM in homologous recombinational repair of DNA damage. EMBO J. 19, 463–471 (2000).
Caspari, T., Murray, J. M. & Carr, A. M. Cdc2-cyclin B kinase activity links Crb2 and Rqh1-topoisomerase III. Genes Dev. 16, 1195–1208 (2002).
Sorensen, C. S. et al. The cell-cycle checkpoint kinase CHK1 is required for mammalian homologous recombination repair. Nature Cell Biol. 7, 195–201 (2005).
Sleeth, K. M. et al. RPA mediates recombination repair during replication stress and is displaced from DNA by checkpoint signalling in human cells. J. Mol. Biol. 373, 38–47 (2007).
Kai, M., Boddy, M. N., Russell, P. & Wang, T. S. Replication checkpoint kinase Cds1 regulates Mus81 to preserve genome integrity during replication stress. Genes Dev. 19, 919–932 (2005).
Taniguchi, T. et al. Convergence of the Fanconi anemia and ataxia telangiectasia signaling pathways. Cell 109, 459–472 (2002).
Niedernhofer, L. J. The Fanconi anemia signalosome anchor. Mol. Cell 25, 487–490 (2007).
Herzberg, K. et al. Phosphorylation of Rad55 on serines 2, 8, and 14 is required for efficient homologous recombination in the recovery of stalled replication forks. Mol. Cell Biol. 26, 8396–8409 (2006).
Flott, S. et al. Phosphorylation of Slx4 by Mec1 and Tel1 regulates the single-strand annealing mode of DNA repair in budding yeast. Mol. Cell Biol. 27, 6433–6445 (2007).
Ziv, Y. et al. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nature Cell Biol. 8, 870–876 (2006).
Stucki, M. & Jackson, S. P. γH2AX and MDC1: anchoring the DNA-damage-response machinery to broken chromosomes. DNA Repair 5, 534–543 (2006).
Kim, S. T., Xu, B. & Kastan, M. B. Involvement of the cohesin protein, Smc1, in Atm-dependent and independent responses to DNA damage. Genes Dev. 16, 560–570 (2002).
Yazdi, P. T. et al. SMC1 is a downstream effector in the ATM/NBS1 branch of the human S-phase checkpoint. Genes Dev. 16, 571–582 (2002).
Lopes, M., Foiani, M. & Sogo, J. M. Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol. Cell 21, 15–27 (2006). Shows physical evidence that TLS and HR mutants accumulate gaps during replication, without affecting fork progression.
Kai, M. & Wang, T. S. Checkpoint activation regulates mutagenic translesion synthesis. Genes Dev. 17, 64–76 (2003). Presents evidence that the 911 damage checkpoint interacts physically with DinB, and promotes DinB loading on chromatin and mutagenic bypass of lesions.
Sabbioneda, S. et al. The 9-1-1 checkpoint clamp physically interacts with polzeta and is partially required for spontaneous polzeta-dependent mutagenesis in Saccharomyces cerevisiae. J. Biol. Chem. 280, 38657–38665 (2005).
Lehmann, A. R. & Fuchs, R. P. Gaps and forks in DNA replication: rediscovering old models. DNA Repair 5, 1495–1498 (2006).
Kai, M., Furuya, K., Paderi, F., Carr, A. M. & Wang, T. S. Rad3-dependent phosphorylation of the checkpoint clamp regulates repair-pathway choice. Nature Cell Biol. 9, 691–697 (2007). Shows that ATM/ATR-dependent phosphorylation of the 911 damage checkpoint promotes Rad6- mediated repair.
Liberi, G. et al. Srs2 DNA helicase is involved in checkpoint response and its regulation requires a functional Mec1-dependent pathway and Cdk1 activity. EMBO J. 19, 5027–5038 (2000).
Ruffner, H., Jiang, W., Craig, A. G., Hunter, T. & Verma, I. M. BRCA1 is phosphorylated at serine 1497 in vivo at a cyclin-dependent kinase 2 phosphorylation site. Mol. Cell Biol. 19, 4843–4854 (1999).
Esashi, F. & Yanagida, M. Cdc2 phosphorylation of Crb2 is required for reestablishing cell cycle progression after the damage checkpoint. Mol. Cell 4, 167–174 (1999).
Moynahan, M. E., Chiu, J. W., Koller, B. H. & Jasin, M. BRCA1 controls homology-directed DNA repair. Mol. Cell 4, 511–518 (1999).
Yu, X., Fu, S., Lai, M., Baer, R. & Chen, J. BRCA1 ubiquitinates its phosphorylation-dependent binding partner CtIP. Genes Dev. 20, 1721–1726 (2006).
Zhao, G. Y. et al. A critical role for the ubiquitin-conjugating enzyme UBC13 in initiating homologous recombination. Mol. Cell 25, 663–675 (2007). Provides the first evidence that the ubiquitin-conjugating activity of UBC13 is required for DSB repair.
Ira, G., Malkova, A., Liberi, G., Foiani, M. & Haber, J. E. Srs2 and Sgs1–Top3 suppress crossovers during double-strand break repair in yeast. Cell 115, 401–411 (2003).
Robert, T., Dervins, D., Fabre, F. & Gangloff, S. Mrc1 and Srs2 are major actors in the regulation of spontaneous crossover. EMBO J. 25, 2837–2846 (2006).
Branzei, D. & Foiani, M. RecQ helicases queuing with Srs2 to disrupt Rad51 filaments and suppress recombination. Genes Dev. 21, 3019–3026 (2007).
Esashi, F. et al. CDK-dependent phosphorylation of BRCA2 as a regulatory mechanism for recombinational repair. Nature 434, 598–604 (2005). Identifies CDK-dependent phosphorylation of BRCA2 and provides evidence that this modification might function as a molecular switch to regulate RAD51 recombination activity.
Yang, H., Li, Q., Fan, J., Holloman, W. K. & Pavletich, N. P. The BRCA2 homologue Brh2 nucleates RAD51 filament formation at a dsDNA–ssDNA junction. Nature 433, 653–657 (2005).
Huen, M. S. et al. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell 131, 901–914 (2007). References 98, 99, 100 and 102 show that the ubiquitin-ligase activity of RNF8 integrates phosphorylation and ubiquitin signalling that is required for DNA repair and checkpoint response.
Kolas, N. K. et al. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science 318, 1637–1640 (2007).
Mailand, N. et al. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell 131, 887–900 (2007).
Plans, V., Guerra-Rebollo, M. & Thomson, T. M. Regulation of mitotic exit by the RNF8 ubiquitin ligase. Oncogene 3 September 2007 (doi:10.1038/sj.onc.1210782).
Wang, B. & Elledge, S. J. Ubc13/Rnf8 ubiquitin ligases control foci formation of the Rap80/Abraxas/Brca1/Brcc36 complex in response to DNA damage. Proc. Natl Acad. Sci. USA 104, 20759–20763 (2007).
Gutierrez, G. J. & Ronai, Z. Ubiquitin and SUMO systems in the regulation of mitotic checkpoints. Trends Biochem. Sci. 31, 324–332 (2006).
Watanabe, N. et al. Cyclin-dependent kinase (CDK) phosphorylation destabilizes somatic Wee1 via multiple pathways. Proc. Natl Acad. Sci. USA 102, 11663–11668 (2005).
Busino, L., Chiesa, M., Draetta, G. F. & Donzelli, M. Cdc25A phosphatase: combinatorial phosphorylation, ubiquitylation and proteolysis. Oncogene 23, 2050–2056 (2004).
Busino, L. et al. Degradation of Cdc25A by β-TrCP during S phase and in response to DNA damage. Nature 426, 87–91 (2003).
Jin, J. et al. SCF-βTRCP links Chk1 signaling to degradation of the Cdc25A protein phosphatase. Genes Dev. 17, 3062–3074 (2003).
Chen, M. S., Ryan, C. E. & Piwnica-Worms, H. Chk1 kinase negatively regulates mitotic function of Cdc25A phosphatase through 14-3-3 binding. Mol. Cell Biol. 23, 7488–7497 (2003).
Smogorzewska, A. et al. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell 129, 289–301 (2007).
Machida, Y. J. et al. UBE2T is the E2 in the Fanconi anemia pathway and undergoes negative autoregulation. Mol. Cell 23, 589–596 (2006).
Meetei, A. R., Yan, Z. & Wang, W. FANCL replaces BRCA1 as the likely ubiquitin ligase responsible for FANCD2 monoubiquitination. Cell Cycle 3, 179–181 (2004).
Andreassen, P. R., D'Andrea, A. D. & Taniguchi, T. ATR couples FANCD2 monoubiquitination to the DNA-damage response. Genes Dev. 18, 1958–1963 (2004).
Ruffner, H., Joazeiro, C. A., Hemmati, D., Hunter, T. & Verma, I. M. Cancer-predisposing mutations within the RING domain of BRCA1: loss of ubiquitin protein ligase activity and protection from radiation hypersensitivity. Proc. Natl Acad. Sci. USA 98, 5134–5139 (2001).
Hashizume, R. et al. The RING heterodimer BRCA1–BARD1 is a ubiquitin ligase inactivated by a breast cancer-derived mutation. J. Biol. Chem. 276, 14537–14540 (2001).
Kim, H., Chen, J. & Yu, X. Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science 316, 1202–1205 (2007).
Wang, B. et al. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science 316, 1194–1198 (2007).
Sobhian, B. et al. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science 316, 1198–1202 (2007).
Stelter, P. & Ulrich, H. D. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature 425, 188–191 (2003). Together with reference 119, this paper provides the first evidence that PCNA monoubiquitylation promotes the TLS damage-tolerance pathway.
Kannouche, P. L., Wing, J. & Lehmann, A. R. Interaction of human DNA polymerase ɛ with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol. Cell 14, 491–500 (2004).
Watanabe, K. et al. Rad18 guides poleta to replication stalling sites through physical interaction and PCNA monoubiquitination. EMBO J. 23, 3886–3896 (2004).
Plosky, B. S. et al. Controlling the subcellular localization of DNA polymerases ι and ɛ via interactions with ubiquitin. EMBO J. 25, 2847–2855 (2006).
Pfander, B., Moldovan, G. L., Sacher, M., Hoege, C. & Jentsch, S. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature 436, 428–433 (2005).
Papouli, E. et al. Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase Srs2p. Mol. Cell 19, 123–133 (2005). References 122 and 123 show that sumoylated PCNA interacts with Srs2 to regulate recombination.
Sacher, M., Pfander, B., Hoege, C. & Jentsch, S. Control of Rad52 recombination activity by double-strand break-induced SUMO modification. Nature Cell Biol. 8, 1284–1290 (2006).
Dieckhoff, P., Bolte, M., Sancak, Y., Braus, G. H. & Irniger, S. Smt3/SUMO and Ubc9 are required for efficient APC/C-mediated proteolysis in budding yeast. Mol. Microbiol. 51, 1375–1387 (2004).
Zhao, X. & Blobel, G. A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc. Natl Acad. Sci. USA 102, 4777–4782 (2005).
Torres-Rosell, J. et al. The Smc5–Smc6 complex and SUMO modification of Rad52 regulates recombinational repair at the ribosomal gene locus. Nature Cell Biol. 9, 923–931 (2007).
Veaute, X. et al. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature 423, 309–312 (2003).
Krejci, L. et al. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature 423, 305–309 (2003).
Bugreev, D. V., Yu, X., Egelman, E. H. & Mazin, A. V. Novel pro- and anti-recombination activities of the Bloom's syndrome helicase. Genes Dev. 21, 3085–3094 (2007).
Hu, Y. et al. RECQL5/Recql5 helicase regulates homologous recombination and suppresses tumor formation via disruption of Rad51 presynaptic filaments. Genes Dev. 21, 3073–3084 (2007).
Liberi, G. et al. Rad51-dependent DNA structures accumulate at damaged replication forks in sgs1 mutants defective in the yeast ortholog of BLM RecQ helicase. Genes Dev. 19, 339–350 (2005). This study and reference 134 provide different types of evidence that the helicase Sgs1 (or BLM in mammals) cooperates with topoisomerase III to dissolve dHJ and pseudo-dHJ, and thus to modulate the outcome of recombination events.
Mankouri, H. W. & Hickson, I. D. Top3 processes recombination intermediates and modulates checkpoint activity after DNA damage. Mol. Biol. Cell 17, 4473–4483 (2006).
Wu, L. & Hickson, I. D. The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature 426, 870–874 (2003).
Branzei, D. et al. Ubc9- and Mms21-mediated sumoylation counteracts recombinogenic events at damaged replication forks. Cell 127, 509–522 (2006). Provides physical evidence that Ubc9- and Mms21-dependent sumoylation functions to prevent recombinogenic structures from accumulating during replication of damaged templates.
Eladad, S. et al. Intra-nuclear trafficking of the BLM helicase to DNA damage-induced foci is regulated by SUMO modification. Hum. Mol. Genet. 14, 1351–1365 (2005).
De Piccoli, G. et al. Smc5–Smc6 mediate DNA double-strand-break repair by promoting sister-chromatid recombination. Nature Cell Biol. 8, 1032–1034 (2006).
Torres-Rosell, J. et al. SMC5 and SMC6 genes are required for the segregation of repetitive chromosome regions. Nature Cell Biol. 7, 412–419 (2005).
Andrews, E. A. et al. Nse2, a component of the Smc5–6 complex, is a SUMO ligase required for the response to DNA damage. Mol. Cell Biol. 25, 185–196 (2005).
Potts, P. R. & Yu, H. Human MMS21/NSE2 is a SUMO ligase required for DNA repair. Mol. Cell Biol. 25, 7021–7032 (2005).
Shiloh, Y. ATM and related protein kinases: safeguarding genome integrity. Nature Rev. Cancer 3, 155–168 (2003).
Kumagai, A. & Dunphy, W. G. Claspin, a novel protein required for the activation of Chk1 during a DNA replication checkpoint response in Xenopus egg extracts. Mol. Cell 6, 839–849 (2000).
Mamely, I. et al. Polo-like kinase-1 controls proteasome-dependent degradation of Claspin during checkpoint recovery. Curr. Biol. 16, 1950–1955 (2006).
Watanabe, N. et al. M-phase kinases induce phospho-dependent ubiquitination of somatic Wee1 by SCF-βTrCP. Proc. Natl Acad. Sci. USA 101, 4419–4424 (2004).
Mailand, N., Bekker-Jensen, S., Bartek, J. & Lukas, J. Destruction of Claspin by SCFβTrCP restrains Chk1 activation and facilitates recovery from genotoxic stress. Mol. Cell 23, 307–318 (2006).
Peschiaroli, A. et al. SCFβTrCP-mediated degradation of Claspin regulates recovery from the DNA replication checkpoint response. Mol. Cell 23, 319–329 (2006).
Geiss-Friedlander, R. & Melchior, F. Concepts in sumoylation: a decade on. Nature Rev. Mol. Cell Biol. 8, 947–956 (2007).
Heun, P. SUMOrganization of the nucleus. Curr. Opin. Cell Biol. 19, 350–355 (2007).
Seeler, J. S., Bischof, O., Nacerddine, K. & Dejean, A. SUMO, the three Rs and cancer. Curr. Top. Microbiol. Immunol. 313, 49–71 (2007).
Burgess, R. C., Rahman, S., Lisby, M., Rothstein, R. & Zhao, X. The Slx5–Slx8 complex affects sumoylation of DNA repair proteins and negatively regulates recombination. Mol. Cell Biol. 27, 6153–6162 (2007).
Xie, Y. et al. The yeast Hex3–Slx8 heterodimer is a ubiquitin ligase stimulated by substrate sumoylation. J. Biol. Chem. 282, 34176–34184 (2007).
Prudden, J. et al. SUMO-targeted ubiquitin ligases in genome stability. EMBO J. 26, 4089–4101 (2007).
Ulrich, H. D. Mutual interactions between the SUMO and ubiquitin systems: a plea of no contest. Trends Cell Biol. 15, 525–532 (2005).
Bylebyl, G. R., Belichenko, I. & Johnson, E. S. The SUMO isopeptidase Ulp2 prevents accumulation of SUMO chains in yeast. J. Biol. Chem. 278, 44113–44120 (2003).
Lopes, M. et al. The DNA replication checkpoint response stabilizes stalled replication forks. Nature 412, 557–561 (2001).
Sogo, J. M., Lopes, M. & Foiani, M. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science 297, 599–602 (2002).
Potts, P. R., Porteus, M. H. & Yu, H. Human SMC5/6 complex promotes sister chromatid homologous recombination by recruiting the SMC1/3 cohesin complex to double-strand breaks. EMBO J. 25, 3377–3388 (2006).
Moldovan, G. L., Pfander, B. & Jentsch, S. PCNA controls establishment of sister chromatid cohesion during S phase. Mol. Cell 23, 723–732 (2006).
Acknowledgements
The authors apologize for the many interesting articles that could not be cited here owing to space limitations. The work in the authors' laboratories is supported by grants form the Associazione Italiana per la Ricerca sul Cancro, the European Community, Telethon-Italy, the Italian Ministry of Education and the Association for International Cancer Research to M.F. and D.B. D.B. is supported by the Buzzati-Traverso foundation.
Author information
Authors and Affiliations
Related links
Glossary
- Replication fork
-
The branch-point structure that forms during DNA replication between the two template DNA strands where nascent DNA synthesis is ongoing.
- Cyclin-dependent kinases
-
A group of serine/threonine protein kinases that are activated at specific points during the cell cycle, together with their regulatory cyclin subunits. They regulate cell-cycle transitions by inducing degradation of cell-cycle inhibitory proteins.
- Apoptosis
-
A form of programmed cell death that is well defined in multicellular organisms.
- Stalled fork
-
A replication fork at which progress is blocked. Progress may be blocked by the presence of bulky lesions, aberrant DNA structures, protein–DNA complexes or depletion of dNTP pools.
- Collapsed forks
-
Disjunction of the two partially replicated sister duplexes at the replication fork that is usually associated with the dissociation of the replisome from the replication fork.
- Replisome
-
Protein machinery that is required to replicate DNA.
- Translesion-synthesis polymerases
-
Low-fidelity and non-processive polymerases that can be used to bypass DNA lesions at the replication fork, often in an error-prone way.
- Template switch
-
(TS). A process that repairs gaps in newly replicated DNA. TS can occur, for example, when a replicative polymerase encounters a lesion on the parental strand. TS uses the information on the newly synthesized sister chromatid as a template to fill in the gaps.
- Topoisomerases
-
Enzymes that remove torsional stress from double-stranded DNA by breaking and rejoining one or two of the DNA strands.
- Supercoils
-
Contortions in DNA that are important for DNA packaging and DNA–RNA synthesis. Topoisomerases sense supercoiling and can either generate or dissipate it by changing DNA topology.
- Precatenanes
-
Cruciform junctions that are formed by the intertwining of the sister duplexes in the replicated portion of a replicone.
- Damage tolerance
-
A post-replicative repair pathway in which the lesions are not repaired, but bypassed (tolerated) during replication. Bypass can be achieved by either using specialized polymerases, or by using the newly synthesized sister chromatid strand as a template.
- Epistasis
-
A group of genes that function in the same biological pathway, usually defined by genetic analysis of double mutants.
- Poly(ADP-ribose) polymerase
-
A polymerase that attaches ADP–ribose moieties to target proteins by means of covalent bonds, which is one of the earliest cellular responses to strand breaks.
- Differentiated cells
-
Cells that are specialized for a particular function (such as neurons and muscle cells) and that cannot proliferate.
- Senescent cells
-
Mitotic cells that cannot divide, but remain metabolically active. Senescence is often caused by stimuli that can cause cancer.
- Double Holliday junction
-
A central intermediate to homologous recombination.
- Ischaemia
-
A restriction in blood supply, generally due to factors in the blood vessels, that causes tissue damage or dysfunction.
- RecQ helicase
-
A family of helicase enzymes that is important for genome maintenance. They function through unwinding paired DNA and translocate in the 3′→5′ direction.
- Fanconi anaemia
-
A rare genetically inherited disorder that is characterized by congenital abnormalities and increased incidence of cancer.
- Srs2
-
A budding yeast DNA helicase that functions to prevent recombination by disrupting Rad51 filaments.
- BRCA1
-
The product of the first breast cancer susceptibility gene; it is involved in DNA repair, cell-cycle regulation and protein ubiquitylation.
- BRCA2
-
A tumour suppressor and an integral component of the homologous-recombination machinery.
- BRCT repeats
-
A protein motif with homology to the C-terminal region of BRCA1 that constitutes a phosphopeptide-recognition domain.
- Hemicatenanes
-
Cruciform junctions of two double-stranded DNA molecules in which one of the strands of one duplex passes between the two strands of the other duplex (and vice versa).
Rights and permissions
About this article
Cite this article
Branzei, D., Foiani, M. Regulation of DNA repair throughout the cell cycle. Nat Rev Mol Cell Biol 9, 297–308 (2008). https://doi.org/10.1038/nrm2351
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrm2351
This article is cited by
-
Uncovering Cell Cycle Dysregulations and Associated Mechanisms in Cancer and Neurodegenerative Disorders: A Glimpse of Hope for Repurposed Drugs
Molecular Neurobiology (2024)
-
Radiotherapy in bone sarcoma: the quest for better treatment option
BMC Cancer (2023)
-
PARP-dependent and NAT10-independent acetylation of N4-cytidine in RNA appears in UV-damaged chromatin
Epigenetics & Chromatin (2023)
-
Targeting of RRM2 suppresses DNA damage response and activates apoptosis in atypical teratoid rhabdoid tumor
Journal of Experimental & Clinical Cancer Research (2023)
-
Functions and regulatory mechanisms of resting hematopoietic stem cells: a promising targeted therapeutic strategy
Stem Cell Research & Therapy (2023)