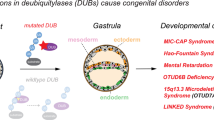
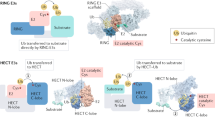
Key Points
-
Ubiquitylation is a post-translational modification that enables mechanistically diverse, quantitative and reversible regulation. Through controlling the stability, interactions or activity of important cellular regulators, ubiquitylation is essential for metazoan development.
-
Aberrant ubiquitylation, most frequently caused by mutation or aberrant expression of genes that encode E3 ubiquitin ligases or deubiquitinases, results in a wide range of developmental diseases, cancer or neurodegeneration.
-
Ubiquitin-dependent protein degradation coordinates proliferation of stem cell populations with the initiation of differentiation and cell fate specification.
-
Ubiquitylation of histone proteins, transcription regulators or ribosome biogenesis factors controls gene expression and mRNA translation programmes that are essential for differentiation.
-
Ubiquitin-dependent regulation of membrane proteins is crucial for cellular communication and cell migration during development.
-
Small molecules that target developmental ubiquitylation enzymes have emerged as a new approach to treating diseases, including cancer.
Abstract
Human development requires intricate cell specification and communication pathways that allow an embryo to generate and appropriately connect more than 200 different cell types. Key to the successful completion of this differentiation programme is the quantitative and reversible regulation of core signalling networks, and post-translational modification with ubiquitin provides embryos with an essential tool to accomplish this task. Instigated by E3 ligases and reversed by deubiquitylases, ubiquitylation controls many processes that are fundamental for development, such as cell division, fate specification and migration. As aberrant function or regulation of ubiquitylation enzymes is at the roots of developmental disorders, cancer, and neurodegeneration, modulating the activity of ubiquitylation enzymes is likely to provide strategies for therapeutic intervention.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Yau, R. & Rape, M. The increasing complexity of the ubiquitin code. Nat. Cell Biol. 18, 579–586 (2016).
Finley, D., Ozkaynak, E. & Varshavsky, A. The yeast polyubiquitin gene is essential for resistance to high temperatures, starvation, and other stresses. Cell 48, 1035–1046 (1987).
Wiborg, O. et al. The human ubiquitin multigene family: some genes contain multiple directly repeated ubiquitin coding sequences. EMBO J. 4, 755–759 (1985).
Finley, D., Bartel, B. & Varshavsky, A. The tails of ubiquitin precursors are ribosomal proteins whose fusion to ubiquitin facilitates ribosome biogenesis. Nature 338, 394–401 (1989).
Ryu, K. Y., Garza, J. C., Lu, X. Y., Barsh, G. S. & Kopito, R. R. Hypothalamic neurodegeneration and adult-onset obesity in mice lacking the Ubb polyubiquitin gene. Proc. Natl Acad. Sci. USA 105, 4016–4021 (2008).
Ryu, H. W., Park, C. W. & Ryu, K. Y. Disruption of polyubiquitin gene Ubb causes dysregulation of neural stem cell differentiation with premature gliogenesis. Sci. Rep. 4, 7026 (2014).
Ryu, K. Y. et al. The mouse polyubiquitin gene UbC is essential for fetal liver development, cell-cycle progression and stress tolerance. EMBO J. 26, 2693–2706 (2007).
Ryu, K. Y., Park, H., Rossi, D. J., Weissman, I. L. & Kopito, R. R. Perturbation of the hematopoietic system during embryonic liver development due to disruption of polyubiquitin gene Ubc in mice. PLoS ONE 7, e32956 (2012).
Chen, X. & Petranovic, D. Role of frameshift ubiquitin B protein in Alzheimer's disease. Wiley Interdiscip. Rev. Syst. Biol. Med. 8, 300–313 (2016).
Lindsten, K. et al. Mutant ubiquitin found in neurodegenerative disorders is a ubiquitin fusion degradation substrate that blocks proteasomal degradation. J. Cell Biol. 157, 417–427 (2002).
Fischer, D. F. et al. Long-term proteasome dysfunction in the mouse brain by expression of aberrant ubiquitin. Neurobiol. Aging 30, 847–863 (2009).
Tank, E. M. & True, H. L. Disease-associated mutant ubiquitin causes proteasomal impairment and enhances the toxicity of protein aggregates. PLoS Genet. 5, e1000382 (2009).
Werner, A. et al. Cell-fate determination by ubiquitin-dependent regulation of translation. Nature 525, 523–527 (2015). This paper demonstrates the control of neural crest specification by non-proteolytic monoubiquitylation of ribosome biogenesis factors.
Jin, L. et al. Ubiquitin-dependent regulation of COPII coat size and function. Nature 482, 495–500 (2012).
Zou, W. et al. The E3 ubiquitin ligase Wwp2 regulates craniofacial development through mono-ubiquitylation of Goosecoid. Nat. Cell Biol. 13, 59–65 (2011).
Chau, V. et al. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science 243, 1576–1583 (1989).
Jin, L., Williamson, A., Banerjee, S., Philipp, I. & Rape, M. Mechanism of ubiquitin-chain formation by the human anaphase-promoting complex. Cell 133, 653–665 (2008).
Meyer, H. J. & Rape, M. Enhanced protein degradation by branched ubiquitin chains. Cell 157, 910–921 (2014).
Wang, C. et al. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature 412, 346–351 (2001).
Spence, J. et al. Cell cycle-regulated modification of the ribosome by a variant multiubiquitin chain. Cell 102, 67–76 (2000).
Tokunaga, F. et al. Involvement of linear polyubiquitylation of NEMO in NF-κB activation. Nat. Cell Biol. 11, 123–132 (2009).
Khaminets, A., Behl, C. & Dikic, I. Ubiquitin-dependent and independent signals in selective autophagy. Trends Cell Biol. 26, 6–16 (2016).
Kane, L. A. et al. PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J. Cell Biol. 205, 143–153 (2014).
Kazlauskaite, A. et al. Parkin is activated by PINK1-dependent phosphorylation of ubiquitin at Ser65. Biochem. J. 460, 127–139 (2014).
Koyano, F. et al. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature 510, 162–166 (2014).
Ohtake, F. et al. Ubiquitin acetylation inhibits polyubiquitin chain elongation. EMBO Rep. 16, 192–201 (2015).
Cui, J. et al. Glutamine deamidation and dysfunction of ubiquitin/NEDD8 induced by a bacterial effector family. Science 329, 1215–1218 (2010).
Bhogaraju, S. et al. Phosphoribosylation of ubiquitin promotes serine ubiquitination and impairs conventional ubiquitination. Cell 167, 1636–1649.e13 (2016).
Schulman, B. A. & Harper, J. W. Ubiquitin-like protein activation by E1 enzymes: the apex for downstream signalling pathways. Nat. Rev. Mol. Cell Biol. 10, 319–331 (2009).
McGrath, J. P., Jentsch, S. & Varshavsky, A. UBA 1: an essential yeast gene encoding ubiquitin-activating enzyme. EMBO J. 10, 227–236 (1991).
Kulkarni, M. & Smith, H. E. E1 ubiquitin-activating enzyme UBA-1 plays multiple roles throughout C. elegans development. PLoS Genet. 4, e1000131 (2008).
Ramser, J. et al. Rare missense and synonymous variants in UBE1 are associated with X-linked infantile spinal muscular atrophy. Am. J. Hum. Genet. 82, 188–193 (2008).
Jin, J., Li, X., Gygi, S. P. & Harper, J. W. Dual E1 activation systems for ubiquitin differentially regulate E2 enzyme charging. Nature 447, 1135–1138 (2007).
Chiu, Y. H., Sun, Q. & Chen, Z. J. E1–L2 activates both ubiquitin and FAT10. Mol. Cell 27, 1014–1023 (2007).
Pelzer, C. et al. UBE1L2, a novel E1 enzyme specific for ubiquitin. J. Biol. Chem. 282, 23010–23014 (2007).
Lee, P. C. et al. Altered social behavior and neuronal development in mice lacking the Uba6–Use1 ubiquitin transfer system. Mol. Cell 50, 172–184 (2013). This study shows how altered ubiquitin activation can affect neuronal development.
Ye, Y. & Rape, M. Building ubiquitin chains: E2 enzymes at work. Nat. Rev. Mol. Cell Biol. 10, 755–764 (2009).
Abdul Rehman, S. A. et al. MINDY-1 is a member of an evolutionarily conserved and structurally distinct new family of deubiquitinating enzymes. Mol. Cell 63, 146–155 (2016).
Sahtoe, D. D. & Sixma, T. K. Layers of DUB regulation. Trends Biochem. Sci. 40, 456–467 (2015).
Komander, D., Clague, M. J. & Urbe, S. Breaking the chains: structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 10, 550–563 (2009).
King, R. W. et al. A 20S complex containing CDC27 and CDC16 catalyzes the mitosis-specific conjugation of ubiquitin to cyclin B. Cell 81, 279–288 (1995).
Zou, H., McGarry, T. J., Bernal, T. & Kirschner, M. W. Identification of a vertebrate sister-chromatid separation inhibitor involved in transformation and tumorigenesis. Science 285, 418–422 (1999).
Li, M., York, J. P. & Zhang, P. Loss of Cdc20 causes a securin-dependent metaphase arrest in two-cell mouse embryos. Mol. Cell. Biol. 27, 3481–3488 (2007).
Visintin, R., Prinz, S. & Amon, A. CDC20 and CDH1: a family of substrate-specific activators of APC-dependent proteolysis. Science 278, 460–463 (1997).
Sivakumar, S. & Gorbsky, G. J. Spatiotemporal regulation of the anaphase-promoting complex in mitosis. Nat. Rev. Mol. Cell Biol. 16, 82–94 (2015).
Eguren, M. et al. The APC/C cofactor Cdh1 prevents replicative stress and p53-dependent cell death in neural progenitors. Nat. Commun. 4, 2880 (2013).
Delgado-Esteban, M., Garcia-Higuera, I., Maestre, C., Moreno, S. & Almeida, A. APC/C-Cdh1 coordinates neurogenesis and cortical size during development. Nat. Commun. 4, 2879 (2013).
Hames, R. S., Wattam, S. L., Yamano, H., Bacchieri, R. & Fry, A. M. APC/C-mediated destruction of the centrosomal kinase Nek2A occurs in early mitosis and depends upon a cyclin A-type D-box. EMBO J. 20, 7117–7127 (2001).
Martins, T., Meghini, F., Florio, F. & Kimata, Y. The APC/C coordinates retinal differentiation with G1 arrest through the Nek2-dependent modulation of Wingless signaling. Dev. Cell 40, 67–80 (2017).
Weber, U. & Mlodzik, M. APC/CFzr/Cdh1-dependent regulation of planar cell polarity establishment via Nek2 kinase acting on Dishevelled. Dev. Cell 40, 53–66 (2017).
Lasorella, A. et al. Degradation of Id2 by the anaphase-promoting complex couples cell cycle exit and axonal growth. Nature 442, 471–474 (2006).
Stegmuller, J. et al. Cell-intrinsic regulation of axonal morphogenesis by the Cdh1–APC target SnoN. Neuron 50, 389–400 (2006).
Yang, Y. et al. A Cdc20–APC ubiquitin signaling pathway regulates presynaptic differentiation. Science 326, 575–578 (2009). References 51–53 demonstrate a role of a cell cycle E3 ligase in differentiation.
Huang, J., Ikeuchi, Y., Malumbres, M. & Bonni, A. A. Cdh1-APC/FMRP ubiquitin signaling link drives mGluR-dependent synaptic plasticity in the mammalian brain. Neuron 86, 726–739 (2015).
Kannan, M., Lee, S. J., Schwedhelm-Domeyer, N. & Stegmuller, J. The E3 ligase Cdh1-anaphase promoting complex operates upstream of the E3 ligase Smurf1 in the control of axon growth. Development 139, 3600–3612 (2012).
van Roessel, P., Elliott, D. A., Robinson, I. M., Prokop, A. & Brand, A. H. Independent regulation of synaptic size and activity by the anaphase-promoting complex. Cell 119, 707–718 (2004).
Fu, A. K. et al. APC(Cdh1) mediates EphA4-dependent downregulation of AMPA receptors in homeostatic plasticity. Nat. Neurosci. 14, 181–189 (2011).
Konishi, Y., Stegmuller, J., Matsuda, T., Bonni, S. & Bonni, A. Cdh1–APC controls axonal growth and patterning in the mammalian brain. Science 303, 1026–1030 (2004).
Juo, P. & Kaplan, J. M. The anaphase-promoting complex regulates the abundance of GLR-1 glutamate receptors in the ventral nerve cord of C. elegans. Curr. Biol. 14, 2057–2062 (2004).
Silies, M. & Klambt, C. APC/C(Fzr/Cdh1)-dependent regulation of cell adhesion controls glial migration in the Drosophila PNS. Nat. Neurosci. 13, 1357–1364 (2010).
Friez, M. J. et al. HUWE1 mutations in Juberg–Marsidi and Brooks syndromes: the results of an X-chromosome exome sequencing study. BMJ Open 6, e009537 (2016).
Urban, N. et al. Return to quiescence of mouse neural stem cells by degradation of a proactivation protein. Science 353, 292–295 (2016). This study shows the important role of the ubiquitin pathway in ensuring proper stem cell division.
King, B. et al. The ubiquitin ligase Huwe1 regulates the maintenance and lymphoid commitment of hematopoietic stem cells. Nat. Immunol. 17, 1312–1321 (2016).
Li, L., Martinez, S. S., Hu, W., Liu, Z. & Tjian, R. A specific E3 ligase/deubiquitinase pair modulates TBP protein levels during muscle differentiation. Elife 4, e08536 (2015).
Forget, A. et al. Shh signaling protects Atoh1 from degradation mediated by the E3 ubiquitin ligase Huwe1 in neural precursors. Dev. Cell 29, 649–661 (2014).
Strohmaier, H. et al. Human F-box protein hCdc4 targets cyclin E for proteolysis and is mutated in a breast cancer cell line. Nature 413, 316–322 (2001).
Koepp, D. M. et al. Phosphorylation-dependent ubiquitination of cyclin E by the SCFFbw7 ubiquitin ligase. Science 294, 173–177 (2001).
Welcker, M. et al. The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc. Natl Acad. Sci. USA 101, 9085–9090 (2004).
Tetzlaff, M. T. et al. Defective cardiovascular development and elevated cyclin E and Notch proteins in mice lacking the Fbw7 F-box protein. Proc. Natl Acad. Sci. USA 101, 3338–3345 (2004).
Reavie, L. et al. Regulation of hematopoietic stem cell differentiation by a single ubiquitin ligase–substrate complex. Nat. Immunol. 11, 207–215 (2010).
Sancho, R., Gruber, R., Gu, G. & Behrens, A. Loss of Fbw7 reprograms adult pancreatic ductal cells into alpha, delta, and beta cells. Cell Stem Cell 15, 139–153 (2014).
Cheng, Y. & Li, G. Role of the ubiquitin ligase Fbw7 in cancer progression. Cancer Metastasis Rev. 31, 75–87 (2012).
Goldknopf, I. L., French, M. F., Musso, R. & Busch, H. Presence of protein A24 in rat liver nucleosomes. Proc. Natl Acad. Sci. USA 74, 5492–5495 (1977).
Gao, Z. et al. PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol. Cell 45, 344–356 (2012).
Levine, S. S. et al. The core of the Polycomb repressive complex is compositionally and functionally conserved in flies and humans. Mol. Cell. Biol. 22, 6070–6078 (2002).
Cao, R., Tsukada, Y. & Zhang, Y. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol. Cell 20, 845–854 (2005).
Wang, H. et al. Role of histone H2A ubiquitination in Polycomb silencing. Nature 431, 873–878 (2004). This study demonstrates the role of histone ubiquitylation in mediating effects of the Polycomb repressive complex.
Endoh, M. et al. Histone H2A mono-ubiquitination is a crucial step to mediate PRC1-dependent repression of developmental genes to maintain ES cell identity. PLoS Genet. 8, e1002774 (2012).
Blackledge, N. P. et al. Variant PRC1 complex-dependent H2A ubiquitylation drives PRC2 recruitment and Polycomb domain formation. Cell 157, 1445–1459 (2014).
Kalb, R. et al. Histone H2A monoubiquitination promotes histone H3 methylation in Polycomb repression. Nat. Struct. Mol. Biol. 21, 569–571 (2014).
Scheuermann, J. C. et al. Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB. Nature 465, 243–247 (2010).
Gu, Y. et al. The histone H2A deubiquitinase Usp16 regulates hematopoiesis and hematopoietic stem cell function. Proc. Natl Acad. Sci. USA 113, E51–E60 (2016).
Jin, J. et al. The deubiquitinase USP21 maintains the stemness of mouse embryonic stem cells via stabilization of Nanog. Nat. Commun. 7, 13594 (2016).
Zhu, P. et al. A histone H2A deubiquitinase complex coordinating histone acetylation and H1 dissociation in transcriptional regulation. Mol. Cell 27, 609–621 (2007).
Wu, X., Johansen, J. V. & Helin, K. Fbxl10/Kdm2b recruits polycomb repressive complex 1 to CpG islands and regulates H2A ubiquitylation. Mol. Cell 49, 1134–1146 (2013).
Voncken, J. W. et al. Rnf2 (Ring1b) deficiency causes gastrulation arrest and cell cycle inhibition. Proc. Natl Acad. Sci. USA 100, 2468–2473 (2003).
del Mar Lorente, M. et al. Loss- and gain-of-function mutations show a polycomb group function for Ring1A in mice. Development 127, 5093–5100 (2000).
van der Lugt, N. M. et al. Posterior transformation, neurological abnormalities, and severe hematopoietic defects in mice with a targeted deletion of the bmi-1 proto-oncogene. Genes Dev. 8, 757–769 (1994).
Li, P. et al. Deubiquitinase MYSM1 is essential for normal bone formation and mesenchymal stem cell differentiation. Sci. Rep. 6, 22211 (2016).
Jiang, X. X. et al. Control of B cell development by the histone H2A deubiquitinase MYSM1. Immunity 35, 883–896 (2011).
Dey, A. et al. Loss of the tumor suppressor BAP1 causes myeloid transformation. Science 337, 1541–1546 (2012). This is an important example showing how aberrant deubiquitylation can cause disease.
Srivastava, A. et al. De novo dominant ASXL3 mutations alter H2A deubiquitination and transcription in Bainbridge–Ropers syndrome. Hum. Mol. Genet. 25, 597–608 (2016).
Jin, J., Arias, E. E., Chen, J., Harper, J. W. & Walter, J. C. A family of diverse Cul4–Ddb1-interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Mol. Cell 23, 709–721 (2006).
Angers, S. et al. Molecular architecture and assembly of the DDB1–CUL4A ubiquitin ligase machinery. Nature 443, 590–593 (2006).
Higa, L. A. et al. CUL4–DDB1 ubiquitin ligase interacts with multiple WD40-repeat proteins and regulates histone methylation. Nat. Cell Biol. 8, 1277–1283 (2006).
Groh, B. S. et al. The antiobesity factor WDTC1 suppresses adipogenesis via the CRL4WDTC1 E3 ligase. EMBO Rep. 17, 638–647 (2016).
Li, G. et al. CRL4DCAF8 ubiquitin ligase targets histone H3K79 and promotes H3K9 methylation in the liver. Cell Rep. 18, 1499–1511 (2017).
Brodersen, M. M. et al. CRL4(WDR23)-mediated SLBP ubiquitylation ensures histone supply during DNA replication. Mol. Cell 62, 627–635 (2016).
Han, J. et al. A Cul4 E3 ubiquitin ligase regulates histone hand-off during nucleosome assembly. Cell 155, 817–829 (2013).
Cang, Y. et al. Deletion of DDB1 in mouse brain and lens leads to p53-dependent elimination of proliferating cells. Cell 127, 929–940 (2006).
Gao, J. et al. The CUL4–DDB1 ubiquitin ligase complex controls adult and embryonic stem cell differentiation and homeostasis. Elife 4, e07539 (2015).
Tarpey, P. S. et al. Mutations in CUL4B, which encodes a ubiquitin E3 ligase subunit, cause an X-linked mental retardation syndrome associated with aggressive outbursts, seizures, relative macrocephaly, central obesity, hypogonadism, pes cavus, and tremor. Am. J. Hum. Genet. 80, 345–352 (2007).
Weintraub, H. et al. The myoD gene family: nodal point during specification of the muscle cell lineage. Science 251, 761–766 (1991).
Takahashi, K. & Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 (2006).
Clevers, H. & Nusse, R. Wnt/β-catenin signaling and disease. Cell 149, 1192–1205 (2012).
Hart, M. et al. The F-box protein β-TrCP associates with phosphorylated β-catenin and regulates its activity in the cell. Curr. Biol. 9, 207–210 (1999).
Latres, E., Chiaur, D. S. & Pagano, M. The human F box protein β-Trcp associates with the Cul1/Skp1 complex and regulates the stability of β-catenin. Oncogene 18, 849–854 (1999).
Winston, J. T. et al. The SCFβ-TRCP-ubiquitin ligase complex associates specifically with phosphorylated destruction motifs in IκBα and β-catenin and stimulates IκBα ubiquitination in vitro. Genes Dev. 13, 270–283 (1999).
Hernandez, A. R., Klein, A. M. & Kirschner, M. W. Kinetic responses of beta-catenin specify the sites of Wnt control. Science 338, 1337–1340 (2012).
Koo, B. K. et al. Tumour suppressor RNF43 is a stem-cell E3 ligase that induces endocytosis of Wnt receptors. Nature 488, 665–669 (2012).
Hao, H. X. et al. ZNRF3 promotes Wnt receptor turnover in an R-spondin-sensitive manner. Nature 485, 195–200 (2012). References 110 and 111 show the role of ubiquitylation in controlling membrane receptors during development.
Pant, V. & Lozano, G. Limiting the power of p53 through the ubiquitin proteasome pathway. Genes Dev. 28, 1739–1751 (2014).
Jaakkola, P. et al. Targeting of HIF-α to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292, 468–472 (2001).
Pfirrmann, T. et al. Hedgehog-dependent E3-ligase Midline1 regulates ubiquitin-mediated proteasomal degradation of Pax6 during visual system development. Proc. Natl Acad. Sci. USA 113, 10103–10108 (2016).
Shimazu, J., Wei, J. & Karsenty, G. Smurf1 inhibits osteoblast differentiation, bone formation, and glucose homeostasis through serine 148. Cell Rep. 15, 27–35 (2016).
Duan, S. et al. FBXO11 targets BCL6 for degradation and is inactivated in diffuse large B-cell lymphomas. Nature 481, 90–93 (2012).
Raducu, M. et al. SCF (Fbxl17) ubiquitylation of Sufu regulates Hedgehog signaling and medulloblastoma development. EMBO J. 35, 1400–1416 (2016).
Quaderi, N. A. et al. Opitz G/BBB syndrome, a defect of midline development, is due to mutations in a new RING finger gene on Xp22. Nat. Genet. 17, 285–291 (1997).
Van Nostrand, J. L. et al. Inappropriate p53 activation during development induces features of CHARGE syndrome. Nature 514, 228–232 (2014).
Kaelin, W. G. Von Hippel-Lindau disease. Annu. Rev. Pathol. 2, 145–173 (2007).
Jia, J. et al. Phosphorylation by double-time/CKIɛ and CKIα targets cubitus interruptus for Slimb/β-TRCP-mediated proteolytic processing. Dev. Cell 9, 819–830 (2005).
Bhatia, N. et al. Gli2 is targeted for ubiquitination and degradation by β-TrCP ubiquitin ligase. J. Biol. Chem. 281, 19320–19326 (2006).
Wang, B. & Li, Y. Evidence for the direct involvement of βTrCP in Gli3 protein processing. Proc. Natl Acad. Sci. USA 103, 33–38 (2006).
Briscoe, J. & Therond, P. P. The mechanisms of Hedgehog signalling and its roles in development and disease. Nat. Rev. Mol. Cell Biol. 14, 416–429 (2013).
Hoppe, T. et al. Activation of a membrane-bound transcription factor by regulated ubiquitin/proteasome-dependent processing. Cell 102, 577–586 (2000).
Palombella, V. J., Rando, O. J., Goldberg, A. L. & Maniatis, T. The ubiquitin–proteasome pathway is required for processing the NF-κB1 precursor protein and the activation of NF-κB. Cell 78, 773–785 (1994).
Signer, R. A., Magee, J. A., Salic, A. & Morrison, S. J. Haematopoietic stem cells require a highly regulated protein synthesis rate. Nature 509, 49–54 (2014).
Blanco, S. et al. Stem cell function and stress response are controlled by protein synthesis. Nature 534, 335–340 (2016).
McCann, K. L. & Baserga, S. J. Genetics. Mysterious ribosomopathies. Science 341, 849–850 (2013).
Sendoel, A. et al. Translation from unconventional 5′ start sites drives tumour initiation. Nature 541, 494–499 (2017).
Lee, A. S., Kranzusch, P. J. & Cate, J. H. eIF3 targets cell-proliferation messenger RNAs for translational activation or repression. Nature 522, 111–114 (2015).
Silva, G. M., Finley, D. & Vogel, C. K63 polyubiquitination is a new modulator of the oxidative stress response. Nat. Struct. Mol. Biol. 22, 116–123 (2015).
Higgins, R. et al. The unfolded protein response triggers site-specific regulatory ubiquitylation of 40S ribosomal proteins. Mol. Cell 59, 35–49 (2015).
Furukawa, M., He, Y. J., Borchers, C. & Xiong, Y. Targeting of protein ubiquitination by BTB–Cullin 3–Roc1 ubiquitin ligases. Nat. Cell Biol. 5, 1001–1007 (2003).
Geyer, R., Wee, S., Anderson, S., Yates, J. & Wolf, D. A. BTB/POZ domain proteins are putative substrate adaptors for cullin 3 ubiquitin ligases. Mol. Cell 12, 783–790 (2003).
Xu, L. et al. BTB proteins are substrate-specific adaptors in an SCF-like modular ubiquitin ligase containing CUL-3. Nature 425, 316–321 (2003).
Dixon, J. et al. Tcof1/Treacle is required for neural crest cell formation and proliferation deficiencies that cause craniofacial abnormalities. Proc. Natl Acad. Sci. USA 103, 13403–13408 (2006).
Hayward, N. K. et al. Whole-genome landscapes of major melanoma subtypes. Nature 545, 175–180 (2017).
McGourty, C. A. et al. Regulation of the CUL3 ubiquitin ligase by a calcium-dependent co-adaptor. Cell 167, 525–538.e14 (2016). This paper provides insight into signalling integration by E3 ligases during development.
Boyadjiev, S. A. et al. Cranio-lenticulo-sutural dysplasia is caused by a SEC23A mutation leading to abnormal endoplasmic-reticulum-to-Golgi trafficking. Nat. Genet. 38, 1192–1197 (2006).
Singer, J. D., Gurian-West, M., Clurman, B. & Roberts, J. M. Cullin-3 targets cyclin E for ubiquitination and controls S phase in mammalian cells. Genes Dev. 13, 2375–2387 (1999).
Lin, G. N. et al. Spatiotemporal 16p11.2 protein network implicates cortical late mid-fetal brain development and KCTD13–Cul3–RhoA pathway in psychiatric diseases. Neuron 85, 742–754 (2015).
O'Roak, B. J. et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature 485, 246–250 (2012).
De Rubeis, S. et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature 515, 209–215 (2014).
Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427 (2014).
Boyden, L. M. et al. Mutations in kelch-like 3 and cullin 3 cause hypertension and electrolyte abnormalities. Nature 482, 98–102 (2012).
Vilchez, D. et al. Increased proteasome activity in human embryonic stem cells is regulated by PSMD11. Nature 489, 304–308 (2012).
Buckley, S. M. et al. Regulation of pluripotency and cellular reprogramming by the ubiquitin–proteasome system. Cell Stem Cell 11, 783–798 (2012).
Brandman, O. & Hegde, R. S. Ribosome-associated protein quality control. Nat. Struct. Mol. Biol. 23, 7–15 (2016).
Bengtson, M. H. & Joazeiro, C. A. Role of a ribosome-associated E3 ubiquitin ligase in protein quality control. Nature 467, 470–473 (2010).
Shao, S., Brown, A., Santhanam, B. & Hegde, R. S. Structure and assembly pathway of the ribosome quality control complex. Mol. Cell 57, 433–444 (2015).
Verma, R., Oania, R. S., Kolawa, N. J. & Deshaies, R. J. Cdc48/p97 promotes degradation of aberrant nascent polypeptides bound to the ribosome. eLife 2, e00308 (2013).
Brandman, O. et al. A ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress. Cell 151, 1042–1054 (2012).
Sundaramoorthy, E. et al. ZNF598 and RACK1 regulate mammalian ribosome-associated quality control function by mediating regulatory 40S ribosomal ubiquitylation. Mol. Cell 65, 751–760.e4 (2017).
Juszkiewicz, S. & Hegde, R. S. Initiation of quality control during Poly(A) translation requires site-specific ribosome ubiquitination. Mol. Cell 65, 743–750.e4 (2017).
Wang, F., Durfee, L. A. & Huibregtse, J. M. A cotranslational ubiquitination pathway for quality control of misfolded proteins. Mol. Cell 50, 368–378 (2013).
Hartl, F. U., Bracher, A. & Hayer-Hartl, M. Molecular chaperones in protein folding and proteostasis. Nature 475, 324–332 (2011).
Chu, J. et al. A mouse forward genetics screen identifies LISTERIN as an E3 ubiquitin ligase involved in neurodegeneration. Proc. Natl Acad. Sci. USA 106, 2097–2103 (2009).
Johnson, J. O. et al. Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron 68, 857–864 (2010).
Kury, S. et al. De novo disruption of the proteasome regulatory subunit PSMD12 causes a syndromic neurodevelopmental disorder. Am. J. Hum. Genet. 100, 352–363 (2017).
Clevers, H., Loh, K. M. & Nusse, R. Stem cell signaling. An integral program for tissue renewal and regeneration: Wnt signaling and stem cell control. Science 346, 1248012 (2014).
Jiang, X., Charlat, O., Zamponi, R., Yang, Y. & Cong, F. Dishevelled promotes Wnt receptor degradation through recruitment of ZNRF3/RNF43 E3 ubiquitin ligases. Mol. Cell 58, 522–533 (2015).
de Lau, W. et al. Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature 476, 293–297 (2011).
Madan, B. et al. USP6 oncogene promotes Wnt signaling by deubiquitylating Frizzleds. Proc. Natl Acad. Sci. USA 113, E2945–E2954 (2016).
Wu, J. et al. Whole-exome sequencing of neoplastic cysts of the pancreas reveals recurrent mutations in components of ubiquitin-dependent pathways. Proc. Natl Acad. Sci. USA 108, 21188–21193 (2011).
Planas-Paz, L. et al. The RSPO–LGR4/5–ZNRF3/RNF43 module controls liver zonation and size. Nat. Cell Biol. 18, 467–479 (2016).
Podos, S. D., Hanson, K. K., Wang, Y. C. & Ferguson, E. L. The DSmurf ubiquitin-protein ligase restricts BMP signaling spatially and temporally during Drosophila embryogenesis. Dev. Cell 1, 567–578 (2001).
Xia, L. et al. The Fused/Smurf complex controls the fate of Drosophila germline stem cells by generating a gradient BMP response. Cell 143, 978–990 (2010).
Persaud, A. et al. Nedd4-1 binds and ubiquitylates activated FGFR1 to control its endocytosis and function. EMBO J. 30, 3259–3273 (2011).
Persaud, A. et al. Tyrosine phosphorylation of NEDD4 activates its ubiquitin ligase activity. Sci. Signal. 7, ra95 (2014).
Sigismund, S. et al. Threshold-controlled ubiquitination of the EGFR directs receptor fate. EMBO J. 32, 2140–2157 (2013).
Paolino, M. et al. The E3 ligase Cbl-b and TAM receptors regulate cancer metastasis via natural killer cells. Nature 507, 508–512 (2014). This is an important study showing how ubiquitin-dependent endocytosis can limit signalling in development and disease.
Yamashita, M. et al. Ubiquitin ligase Smurf1 controls osteoblast activity and bone homeostasis by targeting MEKK2 for degradation. Cell 121, 101–113 (2005).
Blank, M. et al. A tumor suppressor function of Smurf2 associated with controlling chromatin landscape and genome stability through RNF20. Nat. Med. 18, 227–234 (2012).
Broix, L. et al. Mutations in the HECT domain of NEDD4L lead to AKT–mTOR pathway deregulation and cause periventricular nodular heterotopia. Nat. Genet. 48, 1349–1358 (2016).
Sanada, M. et al. Gain-of-function of mutated C-CBL tumour suppressor in myeloid neoplasms. Nature 460, 904–908 (2009).
Yue, S. et al. Requirement of Smurf-mediated endocytosis of Patched1 in sonic hedgehog signal reception. Elife 3, e02555 (2014).
Aster, J. C., Pear, W. S. & Blacklow, S. C. The varied roles of Notch in cancer. Annu. Rev. Pathol. 12, 245–275 (2017).
Pierfelice, T., Alberi, L. & Gaiano, N. Notch in the vertebrate nervous system: an old dog with new tricks. Neuron 69, 840–855 (2011).
Gordon, W. R. et al. Structural basis for autoinhibition of Notch. Nat. Struct. Mol. Biol. 14, 295–300 (2007).
Carrieri, F. A. & Dale, J. K. Turn it down a Notch. Front. Cell Dev. Biol. 4, 151 (2016).
McMillan, B. J. et al. A tail of two sites: a bipartite mechanism for recognition of notch ligands by mind bomb E3 ligases. Mol. Cell 57, 912–924 (2015).
Gordon, W. R. et al. Mechanical allostery: evidence for a force requirement in the proteolytic activation of Notch. Dev. Cell 33, 729–736 (2015). References 182 and 183 provide interesting insight into the role of ubiquitin in cellular communication.
Itoh, M. et al. Mind bomb is a ubiquitin ligase that is essential for efficient activation of Notch signaling by Delta. Dev. Cell 4, 67–82 (2003).
Lai, E. C., Deblandre, G. A., Kintner, C. & Rubin, G. M. Drosophila neuralized is a ubiquitin ligase that promotes the internalization and degradation of delta. Dev. Cell 1, 783–794 (2001).
Koo, B. K. et al. Mind bomb 1 is essential for generating functional Notch ligands to activate Notch. Development 132, 3459–3470 (2005).
Luxan, G. et al. Mutations in the NOTCH pathway regulator MIB1 cause left ventricular noncompaction cardiomyopathy. Nat. Med. 19, 193–201 (2013).
Betancur, P., Bronner-Fraser, M. & Sauka-Spengler, T. Assembling neural crest regulatory circuits into a gene regulatory network. Annu. Rev. Cell Dev. Biol. 26, 581–603 (2010).
Kerosuo, L. & Bronner-Fraser, M. What is bad in cancer is good in the embryo: importance of EMT in neural crest development. Semin. Cell Dev. Biol. 23, 320–332 (2012).
Lander, R., Nordin, K. & LaBonne, C. The F-box protein Ppa is a common regulator of core EMT factors Twist, Snail, Slug, and Sip1. J. Cell Biol. 194, 17–25 (2011).
Campbell, D. S. & Holt, C. E. Chemotropic responses of retinal growth cones mediated by rapid local protein synthesis and degradation. Neuron 32, 1013–1026 (2001).
Deglincerti, A. et al. Coupled local translation and degradation regulate growth cone collapse. Nat. Commun. 6, 6888 (2015).
Menon, S. et al. The E3 ubiquitin ligase TRIM9 is a filopodia off switch required for netrin-dependent axon guidance. Dev. Cell 35, 698–712 (2015).
Winkle, C. C. et al. Trim9 deletion alters the morphogenesis of developing and adult-born hippocampal neurons and impairs spatial learning and memory. J. Neurosci. 36, 4940–4958 (2016).
Hall, J. M. et al. Linkage of early-onset familial breast cancer to chromosome 17q21. Science 250, 1684–1689 (1990).
Long, D. T., Joukov, V., Budzowska, M. & Walter, J. C. BRCA1 promotes unloading of the CMG helicase from a stalled DNA replication fork. Mol. Cell 56, 174–185 (2014).
Reid, L. J. et al. E3 ligase activity of BRCA1 is not essential for mammalian cell viability or homology-directed repair of double-strand DNA breaks. Proc. Natl Acad. Sci. USA 105, 20876–20881 (2008).
Buiting, K., Williams, C. & Horsthemke, B. Angelman syndrome — insights into a rare neurogenetic disorder. Nat. Rev. Neurol. 12, 584–593 (2016).
Yi, J. J. et al. An autism-linked mutation disables phosphorylation control of UBE3A. Cell 162, 795–807 (2015).
Scheffner, M., Huibregtse, J. M., Vierstra, R. D. & Howley, P. M. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell 75, 495–505 (1993).
Kim, H. C. & Huibregtse, J. M. Polyubiquitination by HECT E3s and the determinants of chain type specificity. Mol. Cell. Biol. 29, 3307–3318 (2009).
Greer, P. L. et al. The Angelman syndrome protein Ube3A regulates synapse development by ubiquitinating arc. Cell 140, 704–716 (2010). This article provides insight into the role of UBE3A in neuronal function.
Ito, T. et al. Identification of a primary target of thalidomide teratogenicity. Science 327, 1345–1350 (2010). This is an important study identifying an E3 ligase as the cellular target of the teratogenic small molecule thalidomide.
Fischer, E. S. et al. Structure of the DDB1–CRBN E3 ubiquitin ligase in complex with thalidomide. Nature 512, 49–53 (2014).
Nguyen, T. V. et al. Glutamine triggers acetylation-dependent degradation of glutamine synthetase via the thalidomide receptor cereblon. Mol. Cell 61, 809–820 (2016).
Matyskiela, M. E. et al. A novel cereblon modulator recruits GSPT1 to the CRL4(CRBN) ubiquitin ligase. Nature 535, 252–257 (2016).
Lu, G. et al. The myeloma drug lenalidomide promotes the cereblon-dependent destruction of Ikaros proteins. Science 343, 305–309 (2014). This study shows how targeting of neo-substrates by an E3 ligase can be exploited for therapeutic benefit.
Kronke, J. et al. Lenalidomide induces ubiquitination and degradation of CK1α in del(5q) MDS. Nature 523, 183–188 (2015).
Han, T. et al. Anticancer sulfonamides target splicing by inducing RBM39 degradation via recruitment to DCAF15. Science 356, eaal3755 (2017).
Winter, G. E. et al. Drug development. Phthalimide conjugation as a strategy for in vivo target protein degradation. Science 348, 1376–1381 (2015).
Rape, M. et al. Mobilization of processed, membrane-tethered SPT23 transcription factor by CDC48(UFD1/NPL4), a ubiquitin-selective chaperone. Cell 107, 667–677 (2001).
Acknowledgements
The author apologizes to all colleagues whose work could not be cited owing to space constraints. The author is grateful to J. Schaletzky and all members of his laboratory for continued discussions, fresh ideas, and comments on this manuscript. The author's work is funded by grants from the National Institute of General Medical Sciences. M.R. is an Investigator with the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
M.R. is cofounder of and consultant to Nurix, a biotech company operating in the ubiquitin space.
Glossary
- Dendritic spines
-
Small protrusions from neuronal dendrites; each protrusion is typically connected to a single axon to receive signalling input.
- WNT signals
-
The secreted glycoprotein WNT is often used as a signal to maintain pluripotency or regulate differentiation outcomes.
- Planar cell polarity
-
Coordinated alignment of cell polarity across a tissue plane.
- Haploinsufficiency
-
A condition when a protein product of both alleles is required for sustaining a normal phenotype.
- RING
-
(Really interesting new gene). A signature domain of the largest class of E3 ligases.
- Embryoid bodies
-
3D aggregates of pluripotent stem cells.
- Opitz syndrome
-
A disease in which premature fusion of the metopic suture leads to a triangular shaped forehead.
- CHARGE syndrome
-
A congenital disease that affects eye, nose and ear development.
- Neural crest
-
A cell population that gives rise to melanocytes, cartilage, bone, smooth muscle, peripheral and enteric neurons, and glia.
- Pseudouridylation
-
Isomerization of the uridine nucleoside in ribosomal RNA.
- Small subunit (SSU) processome
-
A ribonucleoprotein complex involved in the processing, maturation and modification of the eukaryotic small ribosomal subunit.
- Spemann organizer
-
A cell cluster in developing amphibian embryos that induces the formation of the central nervous system.
- Paneth cells
-
Cell type in the stem cell niche of the small intestine.
- Intestinal crypts
-
Region at the base of the intestinal epithelium that harbours the stem cells of this organ.
- R-spondin proteins
-
Secreted WNT agonists.
- Natural killer cells
-
Type of lymphocytes in the innate immune system.
- Lateral inhibition
-
The ability of one cell to change the fate or inhibit differentiation of its neighbours.
- Somitogenesis
-
Process of segment formation along the anterior–posterior axis of a developing embryo.
- Epithelial–mesenchymal transition
-
Process by which epithelial cells lose their polarity and cell adhesion and gain migrational and mesenchymal properties.
- Growth cones
-
Dynamic extensions at the tip of a growing axon.
- Dentate gyrus
-
Part of the hippocampus that contributes to memory formation.
- Homologous recombination
-
Genetic recombination between similar DNA molecules, often between sister chromatids during DNA damage repair.
- DNA crosslink
-
A type of DNA damage, with nucleotides becoming covalently linked to each other.
Rights and permissions
About this article
Cite this article
Rape, M. Ubiquitylation at the crossroads of development and disease. Nat Rev Mol Cell Biol 19, 59–70 (2018). https://doi.org/10.1038/nrm.2017.83
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrm.2017.83
This article is cited by
-
Phosphorylation: new star of pathogenesis and treatment in steatotic liver disease
Lipids in Health and Disease (2024)
-
Mechanism of millisecond Lys48-linked poly-ubiquitin chain formation by cullin-RING ligases
Nature Structural & Molecular Biology (2024)
-
Molecular basis of SAP05-mediated ubiquitin-independent proteasomal degradation of transcription factors
Nature Communications (2024)
-
Selenium chemistry for spatio-selective peptide and protein functionalization
Nature Reviews Chemistry (2024)
-
1,10-phenanthroline inhibits sumoylation and reveals that yeast SUMO modifications are highly transient
EMBO Reports (2024)