Abstract
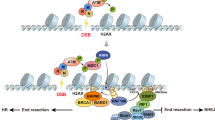
The idea that signal-dependent transcription might involve the generation of transient DNA nicks or even breaks in the regulatory regions of genes, accompanied by activation of DNA damage repair pathways, would seem to be counterintuitive, as DNA damage is usually considered harmful to cellular integrity. However, recent studies have generated a substantial body of evidence that now argues that programmed DNA single- or double-strand breaks can, at least in specific cases, have a role in transcription regulation. Here, we discuss the emerging functions of DNA breaks in the relief of DNA torsional stress and in promoter and enhancer activation.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Wang, J. C. Untangling the Double Helix: DNA Entanglement and the Action of DNA Topoisomerases (Cold Spring Harbor Laboratory Press, 2009).
Pommier, Y., Leo, E., Zhang, H. & Marchand, C. DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem. Biol. 17, 421–433 (2010).
Liu, L. F. & Wang, J. C. Supercoiling of the DNA template during transcription. Proc. Natl Acad. Sci. USA 84, 7024–7027 (1987).
Ma, J. & Wang, M. Interplay between DNA supercoiling and transcription elongation. Transcription 5, e28636 (2014).
Kouzine, F. et al. Transcription-dependent dynamic supercoiling is a short-range genomic force. Nat. Struct. Mol. Biol. 20, 396–403 (2013).
Nelson, P. Transport of torsional stress in DNA. Proc. Natl Acad. Sci. USA 96, 14342–14347 (1999).
Dunaway, M. & Ostrander, E. A. Local domains of supercoiling activate a eukaryotic promoter in vivo. Nature 361, 746–748 (1993).
Parvin, J. D. & Sharp, P. A. DNA topology and a minimal set of basal factors for transcription by RNA polymerase II. Cell 73, 533–540 (1993).
El Hage, A., French, S. L., Beyer, A. L. & Tollervey, D. Loss of topoisomerase I leads to R-loop-mediated transcriptional blocks during ribosomal RNA synthesis. Genes Dev. 24, 1546–1558 (2010).
Wang, J. C. Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell Biol. 3, 430–440 (2002).
Champoux, J. J. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 70, 369–413 (2001).
Gale, K. C. & Osheroff, N. Intrinsic intermolecular DNA ligation activity of eukaryotic topoisomerase II. Potential roles in recombination. J. Biol. Chem. 267, 12090–12097 (1992).
Pommier, Y., Pourquier, P., Fan, Y. & Strumberg, D. Mechanism of action of eukaryotic DNA topoisomerase I and drugs targeted to the enzyme. Biochim. Biophys. Acta 1400, 83–105 (1998).
Pommier, Y., Sun, Y., Huang, S. N. & Nitiss, J. L. Roles of eukaryotic topoisomerases in transcription, replication and genomic stability. Nat. Rev. Mol. Cell Biol. 17, 703–721 (2016).
Vos, S. M., Tretter, E. M., Schmidt, B. H. & Berger, J. M. All tangled up: how cells direct, manage and exploit topoisomerase function. Nat. Rev. Mol. Cell Biol. 12, 827–841 (2011).
Dykhuizen, E. C. et al. BAF complexes facilitate decatenation of DNA by topoisomerase IIα. Nature 497, 624–627 (2013).
Salceda, J., Fernandez, X. & Roca, J. Topoisomerase II, not topoisomerase I, is the proficient relaxase of nucleosomal DNA. EMBO J. 25, 2575–2583 (2006).
Pedersen, J. M. et al. DNA topoisomerases maintain promoters in a state competent for transcriptional activation in Saccharomyces cerevisiae. PLoS Genet. 8, e1003128 (2012).
Kim, T.-K. et al. Widespread transcription at neuronal activity-regulated enhancers. Nature 465, 182–187 (2010).
Corless, S. & Gilbert, N. Effects of DNA supercoiling on chromatin architecture. Biophys. Rev. 8, 245–258 (2016).
Herendeen, D. R., Kassavetis, G. A. & Geiduschek, E. P. A transcriptional enhancer whose function imposes a requirement that proteins track along DNA. Science 256, 1298–1303 (1992).
Roy, D., Zhang, Z., Lu, Z., Hsieh, C. L. & Lieber, M. R. Competition between the RNA transcript and the nontemplate DNA strand during R-loop formation in vitro: a nick can serve as a strong R-loop initiation site. Mol. Cell. Biol. 30, 146–159 (2010).
Belotserkovskii, B. P. et al. Transcription blockage by homopurine DNA sequences: role of sequence composition and single-strand breaks. Nucleic Acids Res. 41, 1817–1828 (2013).
Kuzminov, A. Single-strand interruptions in replicating chromosomes cause double-strand breaks. Proc. Natl Acad. Sci. USA 98, 8241–8246 (2001).
Wimberly, H. et al. R-Loops and nicks initiate DNA breakage and genome instability in non-growing Escherichia coli. Nat. Commun. 4, 2115 (2013).
Ju, B. G. et al. A topoisomerase IIβ-mediated dsDNA break required for regulated transcription. Science 312, 1798–1802 (2006).
Williamson, L. M. & Lees-Miller, S. P. Estrogen receptor α-mediated transcription induces cell cycle-dependent DNA double-strand breaks. Carcinogenesis 32, 279–285 (2011).
Haffner, M. C. et al. Androgen-induced TOP2B-mediated double-strand breaks and prostate cancer gene rearrangements. Nat. Genet. 42, 668–675 (2010).
Lisby, M., Krogh, B. O., Boege, F., Westergaard, O. & Knudsen, B. R. Camptothecins inhibit the utilization of hydrogen peroxide in the ligation step of topoisomerase I catalysis. Biochemistry 37, 10815–10827 (1998).
Ashour, M. E., Atteya, R. & El-Khamisy, S. F. Topoisomerase-mediated chromosomal break repair: an emerging player in many games. Nat. Rev. Cancer 15, 137–151 (2015).
Perillo, B. et al. DNA oxidation as triggered by H3K9me2 demethylation drives estrogen-induced gene expression. Science 319, 202–206 (2008).
Trotter, K. W., King, H. A. & Archer, T. K. Glucocorticoid receptor transcriptional activation via the BRG1-dependent recruitment of TOP2β and Ku70/86. Mol. Cell. Biol. 35, 2799–2817 (2015).
Bunch, H. et al. Transcriptional elongation requires DNA break-induced signalling. Nat. Commun. 6, 10191 (2015).
Baranello, L. et al. RNA polymerase II regulates topoisomerase 1 activity to favor efficient transcription. Cell 165, 357–371 (2016).
Madabhushi, R. et al. Activity-induced DNA breaks govern the expression of neuronal early-response genes. Cell 161, 1592–1605 (2015).
Schwer, B. et al. Transcription-associated processes cause DNA double-strand breaks and translocations in neural stem/progenitor cells. Proc. Natl Acad. Sci. USA 113, 2258–2263 (2016).
Wei, P. C. et al. Long neural genes harbor recurrent DNA break clusters in neural stem/progenitor cells. Cell 164, 644–655 (2016).
King, I. F. et al. Topoisomerases facilitate transcription of long genes linked to autism. Nature 501, 58–62 (2013).
Tiwari, V. K. et al. Target genes of topoisomerase IIβ regulate neuronal survival and are defined by their chromatin state. Proc. Natl Acad. Sci. USA 109, E934–E943 (2012).
Rialdi, A. et al. Topoisomerase 1 inhibition suppresses inflammatory genes and protects from death by inflammation. Science 352, aad7993 (2016).
Puc, J. et al. Ligand-dependent enhancer activation regulated by topoisomerase-I activity. Cell 160, 367–380 (2015).
Bowen, C. et al. NKX3.1 homeodomain protein binds to topoisomerase I and enhances its activity. Cancer Res. 67, 455–464 (2007).
Bowen, C. & Gelmann, E. P. NKX3.1 activates cellular response to DNA damage. Cancer Res. 70, 3089–3097 (2010).
Erbaykent-Tepedelen, B. et al. NKX3.1 contributes to S phase entry and regulates DNA damage response (DDR) in prostate cancer cell lines. Biochem. Biophys. Res. Commun. 414, 123–128 (2011).
Mayeur, G. L. et al. Ku is a novel transcriptional recycling coactivator of the androgen receptor in prostate cancer cells. J. Biol. Chem. 280, 10827–10833 (2005).
Maldonado, E. et al. A human RNA polymerase II complex associated with SRB and DNA-repair proteins. Nature 381, 86–89 (1996).
Stracker, T. H. & Petrini, J. H. The MRE11 complex: starting from the ends. Nat. Rev. Mol. Cell Biol. 12, 90–103 (2011).
Price, B. D. & D'Andrea, A. D. Chromatin remodeling at DNA double-strand breaks. Cell 152, 1344–1354 (2013).
Hartsuiker, E., Neale, M. J. & Carr, A. M. Distinct requirements for the Rad32(Mre11) nuclease and Ctp1(CtIP) in the removal of covalently bound topoisomerase I and II from DNA. Mol. Cell 33, 117–123 (2009).
Hamilton, N. K. & Maizels, N. MRE11 function in response to topoisomerase poisons is independent of its function in double-strand break repair in Saccharomyces cerevisiae. PLoS ONE 5, e15387 (2010).
Sacho, E. J. & Maizels, N. DNA repair factor MRE11/RAD50 cleaves 3′-phosphotyrosyl bonds and resects DNA to repair damage caused by topoisomerase 1 poisons. J. Biol. Chem. 286, 44945–44951 (2011).
Hoa, N. N. et al. Mre11 is essential for the removal of lethal topoisomerase 2 covalent cleavage complexes. Mol. Cell 64, 580–592 (2016).
Mani, R. S. et al. Induced chromosomal proximity and gene fusions in prostate cancer. Science 326, 1230 (2009).
Lin, C. et al. Nuclear receptor-induced chromosomal proximity and DNA breaks underlie specific translocations in cancer. Cell 139, 1069–1083 (2009).
Bansal, K., Yoshida, H., Benoist, C. & Mathis, D. The transcriptional regulator Aire binds to and activates super-enhancers. Nat. Immunol. 18, 263–273 (2017).
Periyasamy, M. et al. APOBEC3B-mediated cytidine deamination is required for estrogen receptor action in breast cancer. Cell Rep. 13, 108–121 (2015).
Qian, J. et al. B cell super-enhancers and regulatory clusters recruit AID tumorigenic activity. Cell 159, 1524–1537 (2014).
Pefanis, E. et al. Noncoding RNA transcription targets AID to divergently transcribed loci in B cells. Nature 514, 389–393 (2014).
Maul, R. W., Saribasak, H., Cao, Z. & Gearhart, P. J. Topoisomerase I deficiency causes RNA polymerase II accumulation and increases AID abundance in immunoglobulin variable genes. DNA Repair (Amst.) 30, 46–52 (2015).
Larsen, B. D. et al. Caspase 3/caspase-activated DNase promote cell differentiation by inducing DNA strand breaks. Proc. Natl Acad. Sci. USA 107, 4230–4235 (2010).
Al-Khalaf, M. H. et al. Temporal activation of XRCC1-mediated DNA repair is essential for muscle differentiation. Cell Discov. 2, 15041 (2016).
Polkinghorn, W. R. et al. Androgen receptor signaling regulates DNA repair in prostate cancers. Cancer Discov. 3, 1245–1253 (2013).
Goodwin, J. F. et al. A hormone-DNA repair circuit governs the response to genotoxic insult. Cancer Discov. 3, 1254–1271 (2013).
Schiewer, M. J. et al. Dual roles of PARP-1 promote cancer growth and progression. Cancer Discov. 2, 1134–1149 (2012).
Krishnakumar, R. & Kraus, W. L. PARP-1 regulates chromatin structure and transcription through a KDM5B-dependent pathway. Mol. Cell 39, 736–749 (2010).
Zhang, Y. et al. Nucleation of DNA repair factors by FOXA1 links DNA demethylation to transcriptional pioneering. Nat. Genet. 48, 1003–1013 (2016).
Zhang, F. et al. Poly(ADP-ribose) polymerase 1 is a key regulator of estrogen receptor α-dependent gene transcription. J. Biol. Chem. 288, 11348–11357 (2013).
Smith, G. C. & Jackson, S. P. The DNA-dependent protein kinase. Genes Dev. 13, 916–934 (1999).
Li, H., Marple, T. & Hasty, P. Ku80-deleted cells are defective at base excision repair. Mutat. Res. 745–746, 16–25 (2013).
Choi, Y. J. et al. Deletion of individual Ku subunits in mice causes an NHEJ-independent phenotype potentially by altering apurinic/apyrimidinic site repair. PLoS ONE 9, e86358 (2014).
Ruscetti, T. et al. Stimulation of the DNA-dependent protein kinase by poly(ADP-ribose) polymerase. J. Biol. Chem. 273, 14461–14467 (1998).
Medunjanin, S. et al. Transcriptional activation of DNA-dependent protein kinase catalytic subunit gene expression by oestrogen receptor-α. EMBO Rep. 11, 208–213 (2010).
Liu, Z. et al. Enhancer activation requires trans-recruitment of a mega transcription factor complex. Cell 159, 358–373 (2014).
Compe, E. & Egly, J. M. TFIIH: when transcription met DNA repair. Nat. Rev. Mol. Cell Biol. 13, 343–354 (2012).
Le May, N., Fradin, D., Iltis, I., Bougneres, P. & Egly, J. M. XPG and XPF endonucleases trigger chromatin looping and DNA demethylation for accurate expression of activated genes. Mol. Cell 47, 622–632 (2012).
Fong, Y. W. et al. A DNA repair complex functions as an Oct4/Sox2 coactivator in embryonic stem cells. Cell 147, 120–131 (2011).
Dixon, J. R. et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380 (2012).
Acknowledgements
The authors apologize to all researchers whose important contributions could not be acknowledged owing to space limitations. The authors thank members of the Rosenfeld laboratory for their comments on the work, and are particularly grateful to P. Cortes and E.P. Geiduschek for discussions. This work was supported by DK 018477, DK 039949, and CA17390. M.G.R. is an Investigator with the Howard Hughes Medical Institute (HHMI).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
PowerPoint slides
Rights and permissions
About this article
Cite this article
Puc, J., Aggarwal, A. & Rosenfeld, M. Physiological functions of programmed DNA breaks in signal-induced transcription. Nat Rev Mol Cell Biol 18, 471–476 (2017). https://doi.org/10.1038/nrm.2017.43
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrm.2017.43
This article is cited by
-
Common occurrence of hotspots of single strand DNA breaks at transcriptional start sites
BMC Genomics (2024)
-
A cyclin D1 intrinsically disordered domain accesses modified histone motifs to govern gene transcription
Oncogenesis (2024)
-
Oxidative stress and ROS-mediated cellular events in RSV infection: potential protective roles of antioxidants
Virology Journal (2023)
-
Complex genomic patterns of abasic sites in mammalian DNA revealed by a high-resolution SSiNGLe-AP method
Nature Communications (2022)
-
Differentiation of human pluripotent stem cells into neurons or cortical organoids requires transcriptional co-regulation by UTX and 53BP1
Nature Neuroscience (2019)