Key Points
-
Cell signalling pathways converge on chromatin, where they induce transcriptional programmes. The inducible programmes are regulated by shared chromatin factors.
-
Targeted inhibition of numerous chromatin factors preferentially suppresses the expression of inducible genes, such as those induced by inflammatory or oncogenic stimuli. Housekeeping genes are not affected by these treatments, as their kinetics of activation and chromatin environment, including the abundance, composition and dependency on some chromatin regulators, are different.
-
Targeting chromatin dependencies unique to the promoter–enhancer structure and composition of inducible genes is an effective, although possibly transient, therapy for cancer and for immunopathologies driven by overt inflammation.
-
The combination of immunotherapy and epigenetic therapy should be carefully tested to evaluate its efficacy. In particular, inhibitors of inducible gene expression programmes might have concurrent effects on cancer cells, immune cells and tumour microenvironment cells.
Abstract
Multiple cell-signalling pathways converge on chromatin to induce gene expression programmes. The inducible transcriptional programmes that are established as a result of inflammatory or oncogenic signals are controlled by shared chromatin regulators. Therapeutic targeting of such chromatin dependencies has proved effective for controlling tumorigenesis and for preventing immunopathologies that are driven by overt inflammation. In this Review, we discuss how chromatin dependencies are established to regulate the expression of key oncogenes and inflammation-promoting genes and how a better mechanistic understanding of such chromatin dependencies can be leveraged to improve the magnitude, timing, duration and selectivity of cell responses with the aim of minimizing unwanted cellular and systemic effects. Recently, exciting progress has been made in cancer immunotherapy and in the development of drugs that target chromatin regulators. We discuss recent advances in clinical trials and the challenge of combining immune-cell-based therapies and epigenetic therapies to improve human health.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Rodbell, M. Nobel Lecture. Signal transduction: evolution of an idea. Biosci. Rep. 15, 117–133 (1995).
Badeaux, A. I. & Shi, Y. Emerging roles for chromatin as a signal integration and storage platform. Nat. Rev. Mol. Cell Biol. 14, 211–224 (2013).
Rialdi, A. et al. Topoisomerase 1 inhibition suppresses inflammatory genes and protects from death by inflammation. Science 352, aad7993 (2016). This paper uncovers a role for TOP1 in the regulation of inducible inflammatory genes and proposes that TOP1 inhibition could be used as therapy for life-threatening infections by dampening overt inflammation.
Baranello, L. et al. RNA polymerase II regulates topoisomerase 1 activity to favor efficient transcription. Cell 165, 357–371 (2016). This article provides mechanistic insights into BRD4-dependent regulation of TOP1 activity, thereby linking transcription elongation and the resolution of DNA topology constraints occurring as a result of transcription.
Nicodeme, E. et al. Suppression of inflammation by a synthetic histone mimic. Nature 468, 1119–1123 (2010).
Clapier, C. R., Iwasa, J., Cairns, B. R. & Peterson, C. L. Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat. Rev. Mol. Cell Biol. 18, 407–422 (2017).
Kadoch, C. et al. Dynamics of BAF-Polycomb complex opposition on heterochromatin in normal and oncogenic states. Nat. Genet. 49, 213–222 (2017).
Stanton, B. Z. et al. Smarca4 ATPase mutations disrupt direct eviction of PRC1 from chromatin. Nat. Genet. 49, 282–288 (2017).
Sif, S., Saurin, A. J., Imbalzano, A. N. & Kingston, R. E. Purification and characterization of mSin3A-containing Brg1 and hBrm chromatin remodeling complexes. Genes Dev. 15, 603–618 (2001).
Gilchrist, D. A. et al. NELF-mediated stalling of Pol II can enhance gene expression by blocking promoter-proximal nucleosome assembly. Genes Dev. 22, 1921–1933 (2008).
Yamaguchi, Y. et al. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell 97, 41–51 (1999).
Kwak, Y. T. et al. Methylation of SPT5 regulates its interaction with RNA polymerase II and transcriptional elongation properties. Mol. Cell 11, 1055–1066 (2003).
Sims, R. J. 3rd et al. The C-terminal domain of RNA polymerase II is modified by site-specific methylation. Science 332, 99–103 (2011).
Zhao, D. Y. et al. SMN and symmetric arginine dimethylation of RNA polymerase II C-terminal domain control termination. Nature 529, 48–53 (2016).
Yang, Y. et al. Arginine methylation facilitates the recruitment of TOP3B to chromatin to prevent R loop accumulation. Mol. Cell 53, 484–497 (2014).
Lin, C. et al. AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol. Cell 37, 429–437 (2010).
Barboric, M., Nissen, R. M., Kanazawa, S., Jabrane-Ferrat, N. & Peterlin, B. M. NF-κB binds P-TEFb to stimulate transcriptional elongation by RNA polymerase II. Mol. Cell 8, 327–337 (2001).
Eberhardy, S. R. & Farnham, P. J. Myc recruits P-TEFb to mediate the final step in the transcriptional activation of the cad promoter. J. Biol. Chem. 277, 40156–40162 (2002).
Galbraith, M. D. et al. HIF1A employs CDK8-mediator to stimulate RNAPII elongation in response to hypoxia. Cell 153, 1327–1339 (2013).
Gardini, A. et al. Integrator regulates transcriptional initiation and pause release following activation. Mol. Cell 56, 128–139 (2014).
Lu, X. et al. Multiple P-TEFbs cooperatively regulate the release of promoter-proximally paused RNA polymerase II. Nucleic Acids Res. 44, 6853–6867 (2016).
Bhagwat, A. S. et al. BET bromodomain inhibition releases the mediator complex from select cis-regulatory elements. Cell Rep. 15, 519–530 (2016).
Jaenicke, L. A. et al. Ubiquitin-Dependent Turnover of MYC antagonizes MYC/PAF1C complex accumulation to drive transcriptional elongation. Mol. Cell 61, 54–67 (2016).
Brown, J. D. et al. NF-κB directs dynamic super enhancer formation in inflammation and atherogenesis. Mol. Cell 56, 219–231 (2014).
Squazzo, S. L. et al. The Paf1 complex physically and functionally associates with transcription elongation factors in vivo. EMBO J. 21, 1764–1774 (2002).
Chen, F. X. et al. PAF1, a molecular regulator of promoter-proximal pausing by RNA polymerase II. Cell 162, 1003–1015 (2015).
Kim, J., Guermah, M. & Roeder, R. G. The human PAF1 complex acts in chromatin transcription elongation both independently and cooperatively with SII/TFIIS. Cell 140, 491–503 (2010).
Marazzi, I. et al. Suppression of the antiviral response by an influenza histone mimic. Nature 483, 428–433 (2012).
Parnas, O. et al. A genome-wide CRISPR screen in primary immune cells to dissect regulatory networks. Cell 162, 675–686 (2015).
Muntean, A. G. et al. The PAF complex synergizes with MLL fusion proteins at HOX loci to promote leukemogenesis. Cancer Cell 17, 609–621 (2010).
Milne, T. A. et al. Multiple interactions recruit MLL1 and MLL1 fusion proteins to the HOXA9 locus in leukemogenesis. Mol. Cell 38, 853–863 (2010).
Baillat, D. et al. Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell 123, 265–276 (2005).
Stadelmayer, B. et al. Integrator complex regulates NELF-mediated RNA polymerase II pause/release and processivity at coding genes. Nat. Commun. 5, 5531 (2014).
Pommier, Y., Sun, Y., Huang, S. N. & Nitiss, J. L. Roles of eukaryotic topoisomerases in transcription, replication and genomic stability. Nat. Rev. Mol. Cell Biol. 17, 703–721 (2016).
Madabhushi, R. et al. Activity-induced DNA breaks govern the expression of neuronal early-response genes. Cell 161, 1592–1605 (2015).
Feinberg, A. P., Koldobskiy, M. A. & Gondor, A. Epigenetic modulators, modifiers and mediators in cancer aetiology and progression. Nat. Rev. Genet. 17, 284–299 (2016).
Kandoth, C. et al. Mutational landscape and significance across 12 major cancer types. Nature 502, 333–339 (2013).
Wajapeyee, N., Malonia, S. K., Palakurthy, R. K. & Green, M. R. Oncogenic RAS directs silencing of tumor suppressor genes through ordered recruitment of transcriptional repressors. Genes Dev. 27, 2221–2226 (2013).
Nabet, B. et al. Deregulation of the Ras-Erk signaling axis modulates the enhancer landscape. Cell Rep. 12, 1300–1313 (2015).
Zippo, A. et al. Histone crosstalk between H3S10ph and H4K16ac generates a histone code that mediates transcription elongation. Cell 138, 1122–1136 (2009).
Wan, L. et al. ENL links histone acetylation to oncogenic gene expression in acute myeloid leukaemia. Nature 543, 265–269 (2017).
Erb, M. A. et al. Transcription control by the ENL YEATS domain in acute leukaemia. Nature 543, 270–274 (2017). Refs 41 and 42 expand our knowledge on the YEATS domain of ENL as an acetyl-lysine histone reader regulating oncogenic transcriptional programmes.
Miller, T. E. et al. Transcription elongation factors represent in vivo cancer dependencies in glioblastoma. Nature 547, 355–359 (2017). This article provides in vivo genetic evidence supporting the key role of chromatin factors in controlling tumorigenesis.
Ray, S. et al. Inducible STAT3 NH2 terminal mono-ubiquitination promotes BRD4 complex formation to regulate apoptosis. Cell Signal. 26, 1445–1455 (2014).
Kadoch, C. et al. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat. Genet. 45, 592–601 (2013).
Shi, J. et al. Role of SWI/SNF in acute leukemia maintenance and enhancer-mediated Myc regulation. Genes Dev. 27, 2648–2662 (2013).
Mathur, R. et al. ARID1A loss impairs enhancer-mediated gene regulation and drives colon cancer in mice. Nat. Genet. 49, 296–302 (2017).
Wang, X. et al. SMARCB1-mediated SWI/SNF complex function is essential for enhancer regulation. Nat. Genet. 49, 289–295 (2017).
Helming, K. C. et al. ARID1B is a specific vulnerability in ARID1A-mutant cancers. Nat. Med. 20, 251–254 (2014).
Kim, K. H. et al. SWI/SNF-mutant cancers depend on catalytic and non-catalytic activity of EZH2. Nat. Med. 21, 1491–1496 (2015).
Wilson, B. G. et al. Epigenetic antagonism between polycomb and SWI/SNF complexes during oncogenic transformation. Cancer Cell 18, 316–328 (2010).
Chovatiya, R. & Medzhitov, R. Stress, inflammation, and defense of homeostasis. Mol. Cell 54, 281–288 (2014).
Delano, M. J. & Ward, P. A. The immune system's role in sepsis progression, resolution, and long-term outcome. Immunol. Rev. 274, 330–353 (2016).
Zhong, Z., Sanchez-Lopez, E. & Karin, M. Autophagy, inflammation, and immunity: a troika governing cancer and its treatment. Cell 166, 288–298 (2016).
Davidson, S., Maini, M. K. & Wack, A. Disease-promoting effects of type I interferons in viral, bacterial, and coinfections. J. Interferon Cytokine Res. 35, 252–264 (2015).
Rodero, M. P. & Crow, Y. J. Type I interferon-mediated monogenic autoinflammation: the type I interferonopathies, a conceptual overview. J. Exp. Med. 213, 2527–2538 (2016).
Ejlerskov, P. et al. Lack of neuronal IFN-β-IFNAR causes Lewy body- and Parkinson's disease-like dementia. Cell 163, 324–339 (2015).
Rice, G. I. et al. Gain-of-function mutations in IFIH1 cause a spectrum of human disease phenotypes associated with upregulated type I interferon signaling. Nat. Genet. 46, 503–509 (2014).
Filippakopoulos, P. et al. Selective inhibition of BET bromodomains. Nature 468, 1067–1073 (2010).
Amit, I. et al. Unbiased reconstruction of a mammalian transcriptional network mediating pathogen responses. Science 326, 257–263 (2009).
Bhatt, D. M. et al. Transcript dynamics of proinflammatory genes revealed by sequence analysis of subcellular RNA fractions. Cell 150, 279–290 (2012).
Iyer, V. R. et al. The transcriptional program in the response of human fibroblasts to serum. Science 283, 83–87 (1999).
Xu, J. et al. Transcriptional competence and the active marking of tissue-specific enhancers by defined transcription factors in embryonic and induced pluripotent stem cells. Genes Dev. 23, 2824–2838 (2009).
Ramirez-Carrozzi, V. R. et al. A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling. Cell 138, 114–128 (2009).
Kininis, M., Isaacs, G. D., Core, L. J., Hah, N. & Kraus, W. L. Postrecruitment regulation of RNA polymerase II directs rapid signaling responses at the promoters of estrogen target genes. Mol. Cell. Biol. 29, 1123–1133 (2009).
Sharma, N., Gabel, H. W. & Greenberg, M. E. A. Shortcut to activity-dependent transcription. Cell 161, 1496–1498 (2015).
Puc, J., Aggarwal, A. K. & Rosenfeld, M. G. Physiological functions of programmed DNA breaks in signal-induced transcription. Nat. Rev. Mol. Cell Biol. 18, 471–476 (2017).
Fernandez, P. C. et al. Genomic targets of the human c-Myc protein. Genes Dev. 17, 1115–1129 (2003).
Guccione, E. et al. Myc-binding-site recognition in the human genome is determined by chromatin context. Nat. Cell Biol. 8, 764–770 (2006).
Sabo, A. et al. Selective transcriptional regulation by Myc in cellular growth control and lymphomagenesis. Nature 511, 488–492 (2014).
Saccani, S., Pantano, S. & Natoli, G. Two waves of nuclear factor κB recruitment to target promoters. J. Exp. Med. 193, 1351–1359 (2001).
Tong, A. J. et al. A stringent systems approach uncovers gene-specific mechanisms regulating inflammation. Cell 165, 165–179 (2016).
Ghisletti, S. et al. Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity 32, 317–328 (2010).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010).
Leung, T. H., Hoffmann, A. & Baltimore, D. One nucleotide in a κB site can determine cofactor specificity for NF-κB dimers. Cell 118, 453–464 (2004).
Thomas, L. R. et al. Interaction with WDR5 promotes target gene recognition and tumorigenesis by MYC. Mol. Cell 58, 440–452 (2015).
Lorenzin, F. et al. Different promoter affinities account for specificity in MYC-dependent gene regulation. eLife 5, e15161 (2016).
Walz, S. et al. Activation and repression by oncogenic MYC shape tumour-specific gene expression profiles. Nature 511, 483–487 (2014).
Guccione, E. et al. Methylation of histone H3R2 by PRMT6 and H3K4 by an MLL complex are mutually exclusive. Nature 449, 933–937 (2007).
Richart, L. et al. BPTF is required for c-MYC transcriptional activity and in vivo tumorigenesis. Nat. Commun. 7, 10153 (2016).
Richart, L., Real, F. X. & Sanchez-Arevalo Lobo, V. J. c-MYC partners with BPTF in human cancer. Mol. Cell. Oncol. 3, e1152346 (2016).
Kress, T. R. et al. Identification of MYC-dependent transcriptional programs in oncogene-addicted liver tumors. Cancer Res. 76, 3463–3472 (2016).
Martinato, F., Cesaroni, M., Amati, B. & Guccione, E. Analysis of Myc-induced histone modifications on target chromatin. PLOS ONE 3, e3650 (2008).
Bouchard, C., Marquardt, J., Bras, A., Medema, R. H. & Eilers, M. Myc-induced proliferation and transformation require Akt-mediated phosphorylation of FoxO proteins. EMBO J. 23, 2830–2840 (2004).
Frank, S. R. et al. MYC recruits the TIP60 histone acetyltransferase complex to chromatin. EMBO Rep. 4, 575–580 (2003).
Liu, X., Tesfai, J., Evrard, Y. A., Dent, S. Y. & Martinez, E. c-Myc transformation domain recruits the human STAGA complex and requires TRRAP and GCN5 acetylase activity for transcription activation. J. Biol. Chem. 278, 20405–20412 (2003).
Pokholok, D. K., Hannett, N. M. & Young, R. A. Exchange of RNA polymerase II initiation and elongation factors during gene expression in vivo. Mol. Cell 9, 799–809 (2002).
Rahl, P. B. et al. c-Myc regulates transcriptional pause release. Cell 141, 432–445 (2010).
Koh, C. M. et al. MYC regulates the core pre-mRNA splicing machinery as an essential step in lymphomagenesis. Nature 523, 96–100 (2015).
Hartono, S. R., Korf, I. F. & Chedin, F. GC skew is a conserved property of unmethylated CpG island promoters across vertebrates. Nucleic Acids Res. 43, 9729–9741 (2015).
Roberts, R. W. & Crothers, D. M. Stability and properties of double and triple helices: dramatic effects of RNA or DNA backbone composition. Science 258, 1463–1466 (1992).
Sanz, L. A. et al. Prevalent, dynamic, and conserved R-loop structures associate with specific epigenomic signatures in mammals. Mol. Cell 63, 167–178 (2016).
Ginno, P. A., Lim, Y. W., Lott, P. L., Korf, I. & Chedin, F. GC skew at the 5′ and 3′ ends of human genes links R-loop formation to epigenetic regulation and transcription termination. Genome Res. 23, 1590–1600 (2013).
Ginno, P. A., Lott, P. L., Christensen, H. C., Korf, I. & Chedin, F. R-Loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol. Cell 45, 814–825 (2012).
Boque-Sastre, R. et al. Head-to-head antisense transcription and R-loop formation promotes transcriptional activation. Proc. Natl Acad. Sci. USA 112, 5785–5790 (2015).
Almada, A. E., Wu, X., Kriz, A. J., Burge, C. B. & Sharp, P. A. Promoter directionality is controlled by U1 snRNP and polyadenylation signals. Nature 499, 360–363 (2013).
Chambers, V. S. et al. High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat. Biotechnol. 33, 877–881 (2015).
Hansel-Hertsch, R. et al. G-Quadruplex structures mark human regulatory chromatin. Nat. Genet. 48, 1267–1272 (2016).
Gonzalez, V. & Hurley, L. H. The C-terminus of nucleolin promotes the formation of the c-MYC G-quadruplex and inhibits c-MYC promoter activity. Biochemistry 49, 9706–9714 (2010).
Li, W., Notani, D. & Rosenfeld, M. G. Enhancers as non-coding RNA transcription units: recent insights and future perspectives. Nat. Rev. Genet. 17, 207–223 (2016).
Aguilo, F. et al. Deposition of 5-methylcytosine on enhancer RNAs enables the coactivator function of PGC-1α. Cell Rep. 14, 479–492 (2016).
Weedon, M. N. et al. Recessive mutations in a distal PTF1A enhancer cause isolated pancreatic agenesis. Nat. Genet. 46, 61–64 (2014).
Maurano, M. T. et al. Systematic localization of common disease-associated variation in regulatory DNA. Science 337, 1190–1195 (2012).
Javierre, B. M. et al. Lineage-specific genome architecture links enhancers and non-coding disease variants to target gene promoters. Cell 167, 1369–1384 (2016).
Lu, Q., Powles, R. L., Wang, Q., He, B. J. & Zhao, H. Integrative tissue-specific functional annotations in the human genome provide novel insights on many complex traits and improve signal prioritization in genome wide association studies. PLoS Genet. 12, e1005947 (2016). Refs 102, 103, 104, 105 provide experimental evidence linking mutations in gene regulatory elements to diseases.
Fulco, C. P. et al. Systematic mapping of functional enhancer-promoter connections with CRISPR interference. Science 354, 769–773 (2016).
Doan, R. N. et al. Mutations in human accelerated regions disrupt cognition and social behavior. Cell 167, 341–354 (2016).
Krijger, P. H. & de Laat, W. Regulation of disease-associated gene expression in the 3D genome. Nat. Rev. Mol. Cell Biol. 17, 771–782 (2016).
Hnisz, D. et al. Activation of proto-oncogenes by disruption of chromosome neighborhoods. Science 351, 1454–1458 (2016).
Donato, E. et al. Compensatory RNA polymerase 2 loading determines the efficacy and transcriptional selectivity of JQ1 in Myc-driven tumors. Leukemia 31, 479–490 (2017).
Chen, C. et al. MLL3 is a haploinsufficient 7q tumor suppressor in acute myeloid leukemia. Cancer Cell 25, 652–665 (2014).
De Raedt, T. et al. PRC2 loss amplifies Ras-driven transcription and confers sensitivity to BRD4-based therapies. Nature 514, 247–251 (2014).
Kieffer-Kwon, K. R. et al. Interactome maps of mouse gene regulatory domains reveal basic principles of transcriptional regulation. Cell 155, 1507–1520 (2013).
Li, G. et al. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell 148, 84–98 (2012).
Dao, L. T. M. et al. Genome-wide characterization of mammalian promoters with distal enhancer functions. Nat. Genet. 49, 1073–1081 (2017).
Farley, E. K., Olson, K. M. & Levine, M. S. Regulatory principles governing tissue specificity of developmental enhancers. Cold Spring Harb. Symp. Quant. Biol. 80, 27–32 (2015).
Peter, I. S. & Davidson, E. H. Evolution of gene regulatory networks controlling body plan development. Cell 144, 970–985 (2011).
Zabidi, M. A. et al. Enhancer-core-promoter specificity separates developmental and housekeeping gene regulation. Nature 518, 556–559 (2015).
Fukaya, T., Lim, B. & Levine, M. Enhancer control of transcriptional bursting. Cell 166, 358–368 (2016). This is a proof-of-principle study showing how an enhancer controls transcription burst frequency and how insulators affect this process.
Senecal, A. et al. Transcription factors modulate c-Fos transcriptional bursts. Cell Rep. 8, 75–83 (2014).
Chuong, E. B., Rumi, M. A., Soares, M. J. & Baker, J. C. Endogenous retroviruses function as species-specific enhancer elements in the placenta. Nat. Genet. 45, 325–329 (2013).
Chuong, E. B., Elde, N. C. & Feschotte, C. Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science 351, 1083–1087 (2016). Refs 121 and 122 show how sequences from human endogenous retroviruses became gene regulatory elements that control gene expression networks.
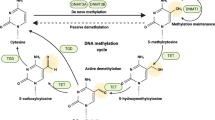
Xiong, J. et al. Cooperative action between SALL4A and TET proteins in stepwise oxidation of 5-methylcytosine. Mol. Cell 64, 913–925 (2016).
Ostuni, R. et al. Latent enhancers activated by stimulation in differentiated cells. Cell 152, 157–171 (2013). This study provides proof of principle for how chromatin and the epigenome can memorize signalling events and deploy latent enhancers to control cell responses in differentiated cells.
Hnisz, D. et al. Super-enhancers in the control of cell identity and disease. Cell 155, 934–947 (2013).
Franke, M. et al. Formation of new chromatin domains determines pathogenicity of genomic duplications. Nature 538, 265–269 (2016).
Groschel, S. et al. A single oncogenic enhancer rearrangement causes concomitant EVI1 and GATA2 deregulation in leukemia. Cell 157, 369–381 (2014).
Affer, M. et al. Promiscuous MYC locus rearrangements hijack enhancers but mostly super-enhancers to dysregulate MYC expression in multiple myeloma. Leukemia 28, 1725–1735 (2014).
Mansour, M. R. et al. Oncogene regulation. An oncogenic super-enhancer formed through somatic mutation of a noncoding intergenic element. Science 346, 1373–1377 (2014).
Heyn, H. et al. Epigenomic analysis detects aberrant super-enhancer DNA methylation in human cancer. Genome Biol. 17, 11 (2016).
Feaver, W. J., Svejstrup, J. Q., Henry, N. L. & Kornberg, R. D. Relationship of CDK-activating kinase and RNA polymerase II CTD kinase TFIIH/TFIIK. Cell 79, 1103–1109 (1994).
Kwiatkowski, N. et al. Targeting transcription regulation in cancer with a covalent CDK7 inhibitor. Nature 511, 616–620 (2014).
Cayrol, F. et al. THZ1 targeting CDK7 suppresses STAT transcriptional activity and sensitizes T-cell lymphomas to BCL2 inhibitors. Nat. Commun. 8, 14290 (2017).
Christensen, C. L. et al. Targeting transcriptional addictions in small cell lung cancer with a covalent CDK7 inhibitor. Cancer Cell 26, 909–922 (2014).
Chipumuro, E. et al. CDK7 inhibition suppresses super-enhancer-linked oncogenic transcription in MYCN-driven cancer. Cell 159, 1126–1139 (2014).
Wang, Y. et al. CDK7-dependent transcriptional addiction in triple-negative breast cancer. Cell 163, 174–186 (2015).
Jiang, Y. Y. et al. Targeting super-enhancer-associated oncogenes in oesophageal squamous cell carcinoma. Gut 66, 1358–1368 (2017).
Loven, J. et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 153, 320–334 (2013).
Lu, H. et al. Compensatory induction of MYC expression by sustained CDK9 inhibition via a BRD4-dependent mechanism. eLife 4, e06535 (2015).
Nilson, K. A. et al. THZ1 reveals roles for Cdk7 in co-transcriptional capping and pausing. Mol. Cell 59, 576–587 (2015).
Titov, D. V. et al. XPB, a subunit of TFIIH, is a target of the natural product triptolide. Nat. Chem. Biol. 7, 182–188 (2011).
Chen, F., Gao, X. & Shilatifard, A. Stably paused genes revealed through inhibition of transcription initiation by the TFIIH inhibitor triptolide. Genes Dev. 29, 39–47 (2015).
Ziaei, S. & Halaby, R. Immunosuppressive, anti-inflammatory and anti-cancer properties of triptolide: a mini review. Avicenna J. Phytomed 6, 149–164 (2016).
Pelish, H. E. et al. Mediator kinase inhibition further activates super-enhancer-associated genes in AML. Nature 526, 273–276 (2015).
Whyte, W. A. et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153, 307–319 (2013).
Kagey, M. H. et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature 467, 430–435 (2010).
Takahashi, H. et al. Human mediator subunit MED26 functions as a docking site for transcription elongation factors. Cell 146, 92–104 (2011).
Zhang, T. et al. Covalent targeting of remote cysteine residues to develop CDK12 and CDK13 inhibitors. Nat. Chem. Biol. 12, 876–884 (2016).
Tyler, D. S. et al. Click chemistry enables preclinical evaluation of targeted epigenetic therapies. Science 356, 1397–1401 (2017). This study presents a strategy based on click chemistry that is used to evaluate the function of chemical compounds; the strategy can be generalized to understand how epigenetic inhibitors function and what their target cells and tissues are.
Zengerle, M., Chan, K. H. & Ciulli, A. Selective small molecule induced degradation of the BET bromodomain protein BRD4. ACS Chem. Biol. 10, 1770–1777 (2015).
Cheung, K. L. et al. Distinct roles of Brd2 and Brd4 in potentiating the transcriptional program for Th17 cell differentiation. Mol. Cell 65, 1068–1080 (2017).
Deeney, J. T., Belkina, A. C., Shirihai, O. S., Corkey, B. E. & Denis, G. V. BET bromodomain proteins Brd2, Brd3 and Brd4 selectively regulate metabolic pathways in the pancreatic β-cell. PLOS ONE 11, e0151329 (2016).
Floyd, S. R. et al. The bromodomain protein Brd4 insulates chromatin from DNA damage signalling. Nature 498, 246–250 (2013).
McKeown, M. R. et al. Biased multicomponent reactions to develop novel bromodomain inhibitors. J. Med. Chem. 57, 9019–9027 (2014).
Sdelci, S. et al. Mapping the chemical chromatin reactivation landscape identifies BRD4-TAF1 cross-talk. Nat. Chem. Biol. 12, 504–510 (2016).
Louder, R. K. et al. Structure of promoter-bound TFIID and model of human pre-initiation complex assembly. Nature 531, 604–609 (2016).
Vangamudi, B. et al. The SMARCA2/4 ATPase domain surpasses the bromodomain as a drug target in SWI/SNF-mutant cancers: insights from cDNA rescue and PFI-3 inhibitor studies. Cancer Res. 75, 3865–3878 (2015).
Wu, Q. et al. Targeting the chromatin remodeling enzyme BRG1 increases the efficacy of chemotherapy drugs in breast cancer cells. Oncotarget 7, 27158–27175 (2016).
Kawano, S. et al. Preclinical evidence of anti-tumor activity induced by EZH2 inhibition in human models of synovial sarcoma. PLoS ONE 11, e0158888 (2016).
Hnisz, D. et al. Convergence of developmental and oncogenic signaling pathways at transcriptional super-enhancers. Mol. Cell 58, 362–370 (2015).
Fong, C. Y. et al. BET inhibitor resistance emerges from leukaemia stem cells. Nature 525, 538–542 (2015).
Rathert, P. et al. Transcriptional plasticity promotes primary and acquired resistance to BET inhibition. Nature 525, 543–547 (2015).
Shu, S. et al. Response and resistance to BET bromodomain inhibitors in triple-negative breast cancer. Nature 529, 413–417 (2016).
Kurimchak, A. M. et al. Resistance to BET bromodomain inhibitors is mediated by kinome reprogramming in ovarian cancer. Cell Rep. 16, 1273–1286 (2016). Refs 139 and 161, 162, 163, 164 highlight the ability of cells to quickly adapt and become resistant to inhibitors of Pol II pause–release factors.
Anand, P. et al. BET bromodomains mediate transcriptional pause release in heart failure. Cell 154, 569–582 (2013).
Felsenstein, K. M. et al. Small molecule microarrays enable the identification of a selective, quadruplex-binding inhibitor of MYC expression. ACS Chem. Biol. 11, 139–148 (2016).
Disney, M. D. et al. A small molecule that targets r(CGG)exp and improves defects in fragile X-associated tremor ataxia syndrome. ACS Chem. Biol. 7, 1711–1718 (2012).
Colak, D. et al. Promoter-bound trinucleotide repeat mRNA drives epigenetic silencing in fragile X syndrome. Science 343, 1002–1005 (2014).
White, R., Saxty, B., Large, J., Kettleborough, C. A. & Jackson, A. P. Identification of small-molecule inhibitors of the ribonuclease H2 enzyme. J. Biomol. Screen. 18, 610–620 (2013).
Kathiravan, M. K., Kale, A. N. & Nilewar, S. Discovery and development of topoisomerase inhibitors as anticancer agents. Mini Rev. Med. Chem. 16, 1219–1229 (2016).
Drolet, M. et al. Overexpression of RNase H partially complements the growth defect of an Escherichia coli ΔtopA mutant: R-loop formation is a major problem in the absence of DNA topoisomerase I. Proc. Natl Acad. Sci. USA 92, 3526–3530 (1995).
Tuduri, S. et al. Topoisomerase I suppresses genomic instability by preventing interference between replication and transcription. Nat. Cell Biol. 11, 1315–1324 (2009).
El Hage, A., French, S. L., Beyer, A. L. & Tollervey, D. Loss of topoisomerase I leads to R-loop-mediated transcriptional blocks during ribosomal RNA synthesis. Genes Dev. 24, 1546–1558 (2010).
Liu, K. et al. Crystal structure of TDRD3 and methyl-arginine binding characterization of TDRD3, SMN and SPF30. PLoS ONE 7, e30375 (2012).
Hu, H., Qian, K., Ho, M. C. & Zheng, Y. G. Small molecule inhibitors of protein arginine methyltransferases. Expert Opin. Investig. Drugs 25, 335–358 (2016).
Skourti-Stathaki, K., Proudfoot, N. J. & Gromak, N. Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol. Cell 42, 794–805 (2011).
Miller, M. S. et al. Senataxin suppresses the antiviral transcriptional response and controls viral biogenesis. Nat. Immunol. 16, 485–494 (2015).
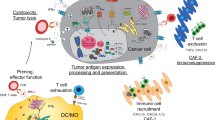
Kourie, H. R. & Klastersky, J. A. Side-effects of checkpoint inhibitor-based combination therapy. Curr. Opin. Oncol. 28, 306–313 (2016).
Benci, J. L. et al. Tumor interferon signaling regulates a multigenic resistance program to immune checkpoint blockade. Cell 167, 1540–1554 (2016).
Utzschneider, D. T. et al. T cells maintain an exhausted phenotype after antigen withdrawal and population reexpansion. Nat. Immunol. 14, 603–610 (2013).
Frebel, H. et al. Programmed death 1 protects from fatal circulatory failure during systemic virus infection of mice. J. Exp. Med. 209, 2485–2499 (2012).
Okazaki, T. et al. PD-1 and LAG-3 inhibitory co-receptors act synergistically to prevent autoimmunity in mice. J. Exp. Med. 208, 395–407 (2011).
Odorizzi, P. M., Pauken, K. E., Paley, M. A., Sharpe, A. & Wherry, E. J. Genetic absence of PD-1 promotes accumulation of terminally differentiated exhausted CD8+ T cells. J. Exp. Med. 212, 1125–1137 (2015).
Sen, D. R. et al. The epigenetic landscape of T cell exhaustion. Science 354, 1165–1169 (2016).
Pauken, K. E. et al. Epigenetic stability of exhausted T cells limits durability of reinvigoration by PD-1 blockade. Science 354, 1160–1165 (2016).
Chiappinelli, K. B. et al. Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell 162, 974–986 (2015).
Roulois, D. et al. DNA-demethylating agents target colorectal cancer cells by inducing viral mimicry by endogenous transcripts. Cell 162, 961–973 (2015).
Ting, D. T. et al. Aberrant overexpression of satellite repeats in pancreatic and other epithelial cancers. Science 331, 593–596 (2011).
Tanne, A. et al. Distinguishing the immunostimulatory properties of noncoding RNAs expressed in cancer cells. Proc. Natl Acad. Sci. USA 112, 15154–15159 (2015). Refs 186, 187, 188, 189 show how the sensing and catabolism of endogenous DNA and RNA can affect tumour progression and therapy.
Goel, S. et al. CDK4/6 inhibition triggers anti-tumour immunity. Nature 548, 471–475 (2017).
Manning, J. et al. Induction of MHC class I molecule cell surface expression and epigenetic activation of antigen-processing machinery components in a murine model for human papilloma virus 16-associated tumours. Immunology 123, 218–227 (2008).
Kim, K. et al. Eradication of metastatic mouse cancers resistant to immune checkpoint blockade by suppression of myeloid-derived cells. Proc. Natl Acad. Sci. USA 111, 11774–11779 (2014).
Chiappinelli, K. B., Zahnow, C. A., Ahuja, N. & Baylin, S. B. Combining epigenetic and immunotherapy to combat cancer. Cancer Res. 76, 1683–1689 (2016).
Kagoya, Y. et al. BET bromodomain inhibition enhances T cell persistence and function in adoptive immunotherapy models. J. Clin. Invest. 126, 3479–3494 (2016).
Chen, R. et al. In vivo RNA interference screens identify regulators of antiviral CD4+ and CD8+ T cell differentiation. Immunity 41, 325–338 (2014).
Banerjee, C. et al. BET bromodomain inhibition as a novel strategy for reactivation of HIV-1. J. Leukoc. Biol. 92, 1147–1154 (2012).
Zhu, H. et al. BET bromodomain inhibition promotes anti-tumor immunity by suppressing PD-L1 expression. Cell Rep. 16, 2829–2837 (2016).
Casey, S. C. et al. MYC regulates the antitumor immune response through CD47 and PD-L1. Science 352, 227–231 (2016).
Hogg, S. J. et al. BET-bromodomain inhibitors engage the host immune system and regulate expression of the immune checkpoint ligand PD-L1. Cell Rep. 18, 2162–2174 (2017). Refs 198 and 199 demonstrate the therapeutic potential of repressing PDL1 expression by targeting regulators of Pol II pause–release and transcription elongation.
Adeegbe, D. O. et al. Synergistic immunostimulatory effects and therapeutic benefit of combined histone deacetylase and bromodomain inhibition in non-small cell lung cancer. Cancer Discov. 7, 852–867 (2017).
Yamamoto, K. et al. Stromal remodeling by the BET bromodomain inhibitor JQ1 suppresses the progression of human pancreatic cancer. Oncotarget 7, 61469–61484 (2016).
Knutson, S. K. et al. Selective inhibition of EZH2 by EPZ-6438 leads to potent antitumor activity in EZH2-mutant non-Hodgkin lymphoma. Mol. Cancer Ther. 13, 842–854 (2014).
Knutson, S. K. et al. Durable tumor regression in genetically altered malignant rhabdoid tumors by inhibition of methyltransferase EZH2. Proc. Natl Acad. Sci. USA 110, 7922–7927 (2013).
Kim, K. H. & Roberts, C. W. Targeting EZH2 in cancer. Nat. Med. 22, 128–134 (2016).
Long, H. et al. The tumor microenvironment disarms CD8+ T lymphocyte function via a miR-26a-EZH2 axis. Oncoimmunology 5, e1245267 (2016).
Shalapour, S. & Karin, M. Immunity, inflammation, and cancer: an eternal fight between good and evil. J. Clin. Invest. 125, 3347–3355 (2015).
Lin, Y., Protter, D. S., Rosen, M. K. & Parker, R. Formation and maturation of phase-separated liquid droplets by RNA-binding proteins. Mol. Cell 60, 208–219 (2015).
Han, T. W. et al. Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell 149, 768–779 (2012).
Hnisz, D., Shrinivas, K., Young, R. A., Chakraborty, A. K. & Sharp, P. A. A. Phase separation model for transcriptional control. Cell 169, 13–23 (2017).
Harlen, K. M. & Churchman, L. S. The code and beyond: transcription regulation by the RNA polymerase II carboxy-terminal domain. Nat. Rev. Mol. Cell Biol. 18, 263–273 (2017).
Albrecht, T. R. & Wagner, E. J. snRNA 3′ end formation requires heterodimeric association of integrator subunits. Mol. Cell. Biol. 32, 1112–1123 (2012).
Yue, J. et al. Integrator orchestrates RAS/ERK1/2 signaling transcriptional programs. Genes Dev. 31, 1809–1820 (2017).
Hubert, C. G. et al. Genome-wide RNAi screens in human brain tumor isolates reveal a novel viability requirement for PHF5A. Genes Dev. 27, 1032–1045 (2013).
Lee, S. C. et al. Modulation of splicing catalysis for therapeutic targeting of leukemia with mutations in genes encoding spliceosomal proteins. Nat. Med. 22, 672–678 (2016).
Gupta, K., Sari-Ak, D., Haffke, M., Trowitzsch, S. & Berger, I. Zooming in on transcription preinitiation. J. Mol. Biol. 428, 2581–2591 (2016).
Giardina, C., Perez-Riba, M. & Lis, J. T. Promoter melting and TFIID complexes on Drosophila genes in vivo. Genes Dev. 6, 2190–2200 (1992).
Rasmussen, E. B. & Lis, J. T. In vivo transcriptional pausing and cap formation on three Drosophila heat shock genes. Proc. Natl Acad. Sci. USA 90, 7923–7927 (1993).
Williams, L. H. et al. Pausing of RNA polymerase II regulates mammalian developmental potential through control of signaling networks. Mol. Cell 58, 311–322 (2015).
Rialdi, A. et al. The RNA exosome syncs IAV-RNAPII transcription to promote viral ribogenesis and infectivity. Cell 169, 679–692 (2017).
Hargreaves, D. C., Horng, T. & Medzhitov, R. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell 138, 129–145 (2009). This paper demonstrates the essential role of promoter-proximal pause–release factors in inducible gene transcription.
Zuber, J. et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature 478, 524–528 (2011). This paper demonstrates the essential role of promoter-proximal pause–release factors in super-enhancer-driven transcription of oncogenes.
La Rosa, P. et al. Sam68 promotes self-renewal and glycolytic metabolism in mouse neural progenitor cells by modulating Aldh1a3 pre-mRNA 3′-end processing. eLife 5, e20750 (2016).
Szczepinska, T. et al. DIS3 shapes the RNA polymerase II transcriptome in humans by degrading a variety of unwanted transcripts. Genome Res. 25, 1622–1633 (2015).
Hazelbaker, D. Z., Marquardt, S., Wlotzka, W. & Buratowski, S. Kinetic competition between RNA Polymerase II and Sen1-dependent transcription termination. Mol. Cell 49, 55–66 (2013).
Sedlyarova, N. et al. sRNA-mediated control of transcription termination in E. coli. Cell 167, 111–121.e13 (2016).
Austenaa, L. M. et al. Transcription of mammalian cis-regulatory elements is restrained by actively enforced early termination. Mol. Cell 60, 460–474 (2015).
Mostafavi, S. et al. Parsing the interferon transcriptional network and its disease associations. Cell 164, 564–578 (2016).
Payne, J. L. & Wagner, A. Mechanisms of mutational robustness in transcriptional regulation. Front. Genet. 6, 322 (2015).
Suter, D. M. et al. Mammalian genes are transcribed with widely different bursting kinetics. Science 332, 472–474 (2011).
Corrigan, A. M., Tunnacliffe, E., Cannon, D. & Chubb, J. R. A continuum model of transcriptional bursting. eLife 5, e13051 (2016).
Weake, V. M. & Workman, J. L. Inducible gene expression: diverse regulatory mechanisms. Nat. Rev. Genet. 11, 426–437 (2010).
Fullwood, M. J. et al. An oestrogen-receptor-α-bound human chromatin interactome. Nature 462, 58–64 (2009).
Daugherty, M. D. & Malik, H. S. Rules of engagement: molecular insights from host-virus arms races. Annu. Rev. Genet. 46, 677–700 (2012).
Marazzi, I. & Garcia-Sastre, A. Interference of viral effector proteins with chromatin, transcription, and the epigenome. Curr. Opin. Microbiol. 26, 123–129 (2015).
Vreede, F. T. & Fodor, E. The role of the influenza virus RNA polymerase in host shut-off. Virulence 1, 436–439 (2010).
Lyles, D. S. Cytopathogenesis and inhibition of host gene expression by RNA viruses. Microbiol. Mol. Biol. Rev. 64, 709–724 (2000).
Browning, D. F. & Busby, S. J. Local and global regulation of transcription initiation in bacteria. Nat. Rev. Microbiol. 14, 638–650 (2016).
Goodrich, J. A. & Tjian, R. Unexpected roles for core promoter recognition factors in cell-type-specific transcription and gene regulation. Nat. Rev. Genet. 11, 549–558 (2010).
Skalska, L., Beltran-Nebot, M., Ule, J. & Jenner, R. G. Regulatory feedback from nascent RNA to chromatin and transcription. Nat. Rev. Mol. Cell Biol. 18, 331–337 (2017).
Acknowledgements
The authors acknowledge that several primary manuscripts could not be duly cited owing to space limitations. The authors thank J. Venables for critical reading and editing of the manuscript. B. Amati, L.. S. Weinberger, S. Campaner, A. Sabo and J. Ho for constructive suggestions and discussions. I.M. thanks D. Charney. The work was partially supported by a seed fund from the Icahn School of Medicine (I.M.), by grant NRF2016-CRP001-103 CRP award (E.G.) and by the RNA Biology Center at the Cancer Science Institute (CSI) of Singapore, National University of Singapore, from funding by the Singapore Ministry of Education's Tier 3 grants, grant number MOE2014-T3-1-006 (D.H.P.L.).
Author information
Authors and Affiliations
Contributions
Researching data for the article: I.M., E.G., B.D.G. and D.H.P.L.; substantial contributions to the discussion of content: I.M. and E.G.; writing: I.M. and E.G.; reviewing and/or editing the manuscript before submission: I.M. and E.G.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information S1 (box)
Cross-comparative analysis of promoter chromatin states with inflammation and oncogenic gene expression signatures. (PDF 5519 kb)
Supplementary information S2 (figure)
Analysis of CpG content or RNA polymerase II (Pol II) occupancy at genes belonging to the categories described in supplementary information S1 (box). (PDF 368 kb)
Supplementary information S3 (box)
Methods used in preparing Figure 3 (PDF 127 kb)
Glossary
- Tonic signals
-
Basal, constitutive and slow-acting signals (in contrast to stimulus-induced and fast-acting signals).
- Chemical probes
-
Small-molecule inhibitors that are potent, selective and cell permeable. They are used in the early stages of drug discovery to study the function of a drug target in a complex biological system.
- Bromodomain and extra-terminal domain
-
(BET). A protein domain of transcription cofactors that allows for interaction with acetylated lysine, mainly on histone tails, to coordinate RNA polymerase II pause–release.
- Inducible gene
-
A gene that is expressed under specific conditions and under the control of a signalling cascade, the expression of which is tightly regulated.
- Immunotherapy
-
The therapeutic use of small molecules, antibodies or cell-based therapies that either stimulate or suppress the immune system or introduce immune system components.
- R-Loops
-
RNA–DNA hybrid structures that can regulate several phases of the transcription process, such as RNA polymerase II processivity and termination.
- Super enhancers
-
Large enhancers with binding sites for multiple transcription factors.
- Pioneer transcription factor
-
A transcription factor that is able to bind compact chromatin and recruit chromatin remodellers, thereby facilitating the subsequent binding of other factors.
- YEATS domain
-
A domain characterized by an immunoglobulin-like fold, which is able to bind specifically to acetylated lysine residues.
- Housekeeping genes
-
Genes that are constitutively expressed in most or all cells of an organism, under a wide range of physiological and pathological conditions. They are essential to maintain cell homeostasis.
- DNA G-quadruplexes
-
Intramolecular and intermolecular four-stranded non-B-DNA structures formed by folding of single-stranded guanine-rich DNA.
- Epigenetic therapy
-
Therapy consisting of small molecules or techniques that target epigenetic regulators of histone post-translational modifications and DNA modifications.
- Immune checkpoints
-
Inhibitory molecules that are essential for modulating the amplitude and duration of immune responses.
- Pattern-recognition receptors
-
Innate immune system receptors present in immune and non-immune cells that recognize pathogen-derived material and induce a protective cellular response.
- Stem cell-like memory T cells
-
T cells at an early and long-lasting stage in the process to becoming memory T cells, between the stages of naive memory T cells and central memory T cells.
- Central memory T cells
-
A long-lived population of memory T cells, which are highly proliferative and produce interleukin-2. They can differentiate into shorter-lived effector memory T cells following antigen stimulation.
- Effector memory T cells
-
A population of mature T cells with low proliferation index and increased levels of interferon-γ and interleukin-4 secretion.
- Adoptive T cell therapy
-
A therapy in which T cells are harvested from a patient and expanded in vitro to enhance their ability to recognize a specific antigen and/or to selectively kill cancer cells. These T cells are then reintroduced into the patient as cell-based therapy.
- T cell receptor-redirected therapy
-
A therapy in which T cells are engineered with an exogenous T cell receptor to redirect antigen specificity.
- Chimeric antigen receptor-redirected therapy
-
A therapy in which T cells are engineered with a chimeric antigen receptor to confer new antigen specificity.
Rights and permissions
About this article
Cite this article
Marazzi, I., Greenbaum, B., Low, D. et al. Chromatin dependencies in cancer and inflammation. Nat Rev Mol Cell Biol 19, 245–261 (2018). https://doi.org/10.1038/nrm.2017.113
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrm.2017.113
This article is cited by
-
Pan-cancer landscape of epigenetic factor expression predicts tumor outcome
Communications Biology (2023)
-
Dependency of NELF-E-SLUG-KAT2B epigenetic axis in breast cancer carcinogenesis
Nature Communications (2023)
-
A novel chromatin regulator-related immune checkpoint related gene prognostic signature and potential candidate drugs for endometrial cancer patients
Hereditas (2022)
-
Identification of a chromatin regulator signature and potential candidate drugs for bladder cancer
Hereditas (2022)
-
BET bromodomain inhibitors regulate keratinocyte plasticity
Nature Chemical Biology (2021)