Key Points
-
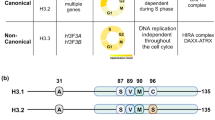
Histone variants have evolved to endow chromatin with special properties in a locus-specific manner, and they differ from replication-coupled ('canonical') histones in terms of their gene composition, RNA processing, expression and deposition timing, and protein structure.
-
Specific chaperones and chromatin remodellers regulate the chromatin incorporation and removal of histone variants, and they can act in a locus-specific manner.
-
Histone variants are dynamically expressed during early embryonic development and have specialized functions during lineage commitment and during somatic cell reprogramming.
-
In cancer, histone variants and their regulators are frequently deregulated at the level of transcription and in some cases by mutations.
-
The deregulation of histone variants contributes to cancer through multiple mechanisms, including altered transcription, increased epigenetic plasticity and the induction of genomic instability.
-
As an example, lysine residues 27 and 36 of H3.3 (as well as those of the replication-coupled H3.1) can be modified by methylation and are frequently mutated in childhood cancers. These are gain-of-function mutations. The mutant proteins act as inhibitors of specific histone methyltransferases and probably drive cancer by perturbing epigenetic regulation in a restricted developmental time window.
Abstract
Histone variants endow chromatin with unique properties and show a specific genomic distribution that is regulated by specific deposition and removal machineries. These variants — in particular, H2A.Z, macroH2A and H3.3 — have important roles in early embryonic development, and they regulate the lineage commitment of stem cells, as well as the converse process of somatic cell reprogramming to pluripotency. Recent progress has also shed light on how mutations, transcriptional deregulation and changes in the deposition machineries of histone variants affect the process of tumorigenesis. These alterations promote or even drive cancer development through mechanisms that involve changes in epigenetic plasticity, genomic stability and senescence, and by activating and sustaining cancer-promoting gene expression programmes.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Talbert, P. B. et al. A unified phylogeny-based nomenclature for histone variants. Epigenetics Chromatin 5, 7 (2012).
Siegel, T. N. et al. Four histone variants mark the boundaries of polycistronic transcription units in Trypanosoma brucei. Genes Dev. 23, 1063–1076 (2009).
Moosmann, A. et al. Histone variant innovation in a rapidly evolving chordate lineage. BMC Evol. Biol. 11, 208 (2011).
Albig, W. & Doenecke, D. The human histone gene cluster at the D6S105 locus. Hum. Genet. 101, 284–294 (1997).
Albig, W., Kioschis, P., Poustka, A., Meergans, K. & Doenecke, D. Human histone gene organization: nonregular arrangement within a large cluster. Genomics 40, 314–322 (1997).
Ivanova, V. S., Zimonjic, D., Popescu, N. & Bonner, W. M. Chromosomal localization of the human histone H2A.X gene to 11q23.2-q23.3 by fluorescence in situ hybridization. Hum. Genet. 94, 303–306 (1994).
Govin, J., Caron, C., Lestrat, C., Rousseaux, S. & Khochbin, S. The role of histones in chromatin remodelling during mammalian spermiogenesis. Eur. J. Biochem. 271, 3459–3469 (2004).
Cakmakci, N. G., Lerner, R. S., Wagner, E. J., Zheng, L. & Marzluff, W. F. SLIP1, a factor required for activation of histone mRNA translation by the stem-loop binding protein. Mol. Cell. Biol. 28, 1182–1194 (2008).
Bonisch, C. & Hake, S. B. Histone H2A variants in nucleosomes and chromatin: more or less stable? Nucleic Acids Res. 40, 10719–10741 (2012).
Rasmussen, T. P. et al. Messenger RNAs encoding mouse histone macroH2A1 isoforms are expressed at similar levels in male and female cells and result from alternative splicing. Nucleic Acids Res. 27, 3685–3689 (1999).
Bonisch, C. et al. H2A.Z.2.2 is an alternatively spliced histone H2A.Z variant that causes severe nucleosome destabilization. Nucleic Acids Res. 40, 5951–5964 (2012).
Marzluff, W. F., Wagner, E. J. & Duronio, R. J. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat. Rev. Genet. 9, 843–854 (2008).
Talbert, P. B. & Henikoff, S. Histone variants — ancient wrap artists of the epigenome. Nat. Rev. Mol. Cell Biol. 11, 264–275 (2010).
Talbert, P. B. & Henikoff, S. Environmental responses mediated by histone variants. Trends Cell Biol. 24, 642–650 (2014).
Skene, P. J. & Henikoff, S. Histone variants in pluripotency and disease. Development 140, 2513–2524 (2013).
Filipescu, D., Muller, S. & Almouzni, G. Histone H3 variants and their chaperones during development and disease: contributing to epigenetic control. Annu. Rev. Cell Dev. Biol. 30, 615–646 (2014).
Gurard-Levin, Z. A., Quivy, J. P. & Almouzni, G. Histone chaperones: assisting histone traffic and nucleosome dynamics. Annu. Rev. Biochem. 83, 487–517 (2014).
Luger, K., Mader, A. W., Richmond, R. K., Sargent, D. F. & Richmond, T. J. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389, 251–260 (1997).
Mattiroli, F., D'Arcy, S. & Luger, K. The right place at the right time: chaperoning core histone variants. EMBO Rep. 16, 1454–1466 (2015).
Burgess, R. J. & Zhang, Z. Histone chaperones in nucleosome assembly and human disease. Nat. Struct. Mol. Biol. 20, 14–22 (2013).
Cook, A. J., Gurard-Levin, Z. A., Vassias, I. & Almouzni, G. A specific function for the histone chaperone NASP to fine-tune a reservoir of soluble H3-H4 in the histone supply chain. Mol. Cell 44, 918–927 (2011).
Campos, E. I. et al. The program for processing newly synthesized histones H3.1 and H4. Nat. Struct. Mol. Biol. 17, 1343–1351 (2010).
Elsasser, S. J. A common structural theme in histone chaperones mimics interhistone contacts. Trends Biochem. Sci. 38, 333–336 (2013).
Nakatani, Y., Ray-Gallet, D., Quivy, J. P., Tagami, H. & Almouzni, G. Two distinct nucleosome assembly pathways: dependent or independent of DNA synthesis promoted by histone H3.1 and H3.3 complexes. Cold Spring Harb. Symp. Quant. Biol. 69, 273–280 (2004).
Narlikar, G. J., Sundaramoorthy, R. & Owen-Hughes, T. Mechanisms and functions of ATP-dependent chromatin-remodeling enzymes. Cell 154, 490–503 (2013).
Goldberg, A. D. et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell 140, 678–691 (2010). In this paper, different chaperone and chromatin-remodelling complexes are shown to control the distribution of H3.3 in euchromatin and heterochromatin.
Ray-Gallet, D. et al. Dynamics of histone H3 deposition in vivo reveal a nucleosome gap-filling mechanism for H3.3 to maintain chromatin integrity. Mol. Cell 44, 928–941 (2011).
Drane, P., Ouararhni, K., Depaux, A., Shuaib, M. & Hamiche, A. The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes Dev. 24, 1253–1265 (2010).
Wong, L. H. et al. Histone H3.3 incorporation provides a unique and functionally essential telomeric chromatin in embryonic stem cells. Genome Res. 19, 404–414 (2009).
Wong, L. H. et al. ATRX interacts with H3.3 in maintaining telomere structural integrity in pluripotent embryonic stem cells. Genome Res. 20, 351–360 (2010).
Elsasser, S. J., Noh, K. M., Diaz, N., Allis, C. D. & Banaszynski, L. A. Histone H3.3 is required for endogenous retroviral element silencing in embryonic stem cells. Nature 522, 240–244 (2015).
Sadic, D. et al. Atrx promotes heterochromatin formation at retrotransposons. EMBO Rep. 16, 836–850 (2015).
Ratnakumar, K. et al. ATRX-mediated chromatin association of histone variant macroH2A1 regulates α-globin expression. Genes Dev. 26, 433–438 (2012).
Billon, P. & Cote, J. Precise deposition of histone H2A.Z in chromatin for genome expression and maintenance. Biochim. Biophys. Acta 1819, 290–302 (2013).
Obri, A. et al. ANP32E is a histone chaperone that removes H2A.Z from chromatin. Nature 505, 648–653 (2014).
Mao, Z. et al. Anp32e, a higher eukaryotic histone chaperone directs preferential recognition for H2A.Z. Cell Res. 24, 389–399 (2014).
Gursoy-Yuzugullu, O., Ayrapetov, M. K. & Price, B. D. Histone chaperone Anp32e removes H2A.Z from DNA double-strand breaks and promotes nucleosome reorganization and DNA repair. Proc. Natl Acad. Sci. USA 112, 7507–7512 (2015).
Bao, Y. et al. Nucleosomes containing the histone variant H2A.Bbd organize only 118 base pairs of DNA. EMBO J. 23, 3314–3324 (2004).
Doyen, C. M. et al. Dissection of the unusual structural and functional properties of the variant H2A.Bbd nucleosome. EMBO J. 25, 4234–4244 (2006).
Zhou, J., Fan, J. Y., Rangasamy, D. & Tremethick, D. J. The nucleosome surface regulates chromatin compaction and couples it with transcriptional repression. Nat. Struct. Mol. Biol. 14, 1070–1076 (2007).
Bui, M., Walkiewicz, M. P., Dimitriadis, E. K. & Dalal, Y. The CENP-A nucleosome: a battle between Dr Jekyll and Mr Hyde. Nucleus 4, 37–42 (2013).
Muller, S. et al. Phosphorylation and DNA binding of HJURP determine its centromeric recruitment and function in CenH3CENP-A loading. Cell Rep. 8, 190–203 (2014).
Bernstein, E. & Hake, S. B. The nucleosome: a little variation goes a long way. Biochem. Cell Biol. 84, 505–517 (2006).
Jin, C. et al. H3.3/H2A.Z double variant-containing nucleosomes mark 'nucleosome-free regions' of active promoters and other regulatory regions. Nat. Genet. 41, 941–945 (2009).
Chen, P., Wang, Y. & Li, G. Dynamics of histone variant H3.3 and its coregulation with H2A.Z at enhancers and promoters. Nucleus 5, 21–27 (2014).
de Dieuleveult, M. et al. Genome-wide nucleosome specificity and function of chromatin remodellers in ES cells. Nature 530, 113–116 (2016).
Arimura, Y. et al. Crystal structure and stable property of the cancer-associated heterotypic nucleosome containing CENP-A and H3.3. Sci. Rep. 4, 7115 (2014).
Lacoste, N. et al. Mislocalization of the centromeric histone variant CenH3/CENP-A in human cells depends on the chaperone DAXX. Mol. Cell 53, 631–644 (2014).
Allis, C. D., Caparras, M. L., Jenuwein, T. & Reinberg, D. (eds) Epigenetics 2nd edn (Cold Spring Harbor Laboratory Press, 2015).
McKittrick, E., Gafken, P. R., Ahmad, K. & Henikoff, S. Histone H3.3 is enriched in covalent modifications associated with active chromatin. Proc. Natl Acad. Sci. USA 101, 1525–1530 (2004).
Hake, S. B. et al. Expression patterns and post-translational modifications associated with mammalian histone H3 variants. J. Biol. Chem. 281, 559–568 (2006).
Singh, N. et al. Dual recognition of phosphoserine and phosphotyrosine in histone variant H2A.X by DNA damage response protein MCPH1. Proc. Natl Acad. Sci. USA 109, 14381–14386 (2012).
Draker, R. et al. A combination of H2A.Z and H4 acetylation recruits Brd2 to chromatin during transcriptional activation. PLoS Genet. 8, e1003047 (2012). This study shows that BRD2 mediates H2A.Z function by recognizing both the histone variant and its post-translational modifications.
Wen, H. et al. ZMYND11 links histone H3.3K36me3 to transcription elongation and tumour suppression. Nature 508, 263–268 (2014). This paper reports that the recognition of K36me3 on histone variant H3.3 by the PWWP-domain protein ZMYND11 has a role in regulating transcription elongation and suppressing tumorigenesis.
Timinszky, G. et al. A macrodomain-containing histone rearranges chromatin upon sensing PARP1 activation. Nat. Struct. Mol. Biol. 16, 923–929 (2009).
Chen, H. et al. MacroH2A1.1 and PARP-1 cooperate to regulate transcription by promoting CBP-mediated H2B acetylation. Nat. Struct. Mol. Biol. 21, 981–989 (2014).
Nashun, B., Yukawa, M., Liu, H., Akiyama, T. & Aoki, F. Changes in the nuclear deposition of histone H2A variants during pre-implantation development in mice. Development 137, 3785–3794 (2010).
Torres-Padilla, M. E., Bannister, A. J., Hurd, P. J., Kouzarides, T. & Zernicka-Goetz, M. Dynamic distribution of the replacement histone variant H3.3 in the mouse oocyte and preimplantation embryos. Int. J. Dev. Biol. 50, 455–461 (2006).
Rangasamy, D., Berven, L., Ridgway, P. & Tremethick, D. J. Pericentric heterochromatin becomes enriched with H2A.Z during early mammalian development. EMBO J. 22, 1599–1607 (2003).
Creppe, C. et al. MacroH2A1 regulates the balance between self-renewal and differentiation commitment in embryonic and adult stem cells. Mol. Cell. Biol. 32, 1442–1452 (2012). This study demonstrates that macroH2A1.2 promotes the lineage commitment of mouse ES cells and that its loss provokes the growth of undifferentiated teratoma tissue.
Buschbeck, M. et al. The histone variant macroH2A is an epigenetic regulator of key developmental genes. Nat. Struct. Mol. Biol. 16, 1074–1079 (2009). This study was the first to demonstrate that macroH2A co-localizes with Polycomb-marked genes and that it is required for robust embryonic development in zebrafish.
Mendiburo, M. J., Padeken, J., Fulop, S., Schepers, A. & Heun, P. Drosophila CENH3 is sufficient for centromere formation. Science 334, 686–690 (2011).
Howman, E. V. et al. Early disruption of centromeric chromatin organization in centromere protein A (Cenpa) null mice. Proc. Natl Acad. Sci. USA 97, 1148–1153 (2000).
Black, B. E. et al. Centromere identity maintained by nucleosomes assembled with histone H3 containing the CENP-A targeting domain. Mol. Cell 25, 309–322 (2007).
Fachinetti, D. et al. A two-step mechanism for epigenetic specification of centromere identity and function. Nat. Cell Biol. 15, 1056–1066 (2013).
Jang, C. W., Shibata, Y., Starmer, J., Yee, D. & Magnuson, T. Histone H3.3 maintains genome integrity during mammalian development. Genes Dev. 29, 1377–1392 (2015).
Couldrey, C., Carlton, M. B., Nolan, P. M., Colledge, W. H. & Evans, M. J. A retroviral gene trap insertion into the histone 3.3A gene causes partial neonatal lethality, stunted growth, neuromuscular deficits and male sub-fertility in transgenic mice. Hum. Mol. Genet. 8, 2489–2495 (1999).
Bush, K. M. et al. Endogenous mammalian histone H3.3 exhibits chromatin-related functions during development. Epigenetics Chromatin 6, 7 (2013).
Tang, M. C. et al. Contribution of the two genes encoding histone variant H3.3 to viability and fertility in mice. PLoS Genet. 11, e1004964 (2015).
Santenard, A. et al. Heterochromatin formation in the mouse embryo requires critical residues of the histone variant H3.3. Nat. Cell Biol. 12, 853–862 (2010).
Rangasamy, D., Greaves, I. & Tremethick, D. J. RNA interference demonstrates a novel role for H2A.Z in chromosome segregation. Nat. Struct. Mol. Biol. 11, 650–655 (2004).
Faast, R. et al. Histone variant H2A.Z is required for early mammalian development. Curr. Biol. 11, 1183–1187 (2001).
Bassing, C. H. et al. Histone H2AX: a dosage-dependent suppressor of oncogenic translocations and tumors. Cell 114, 359–370 (2003).
Celeste, A. et al. Genomic instability in mice lacking histone H2AX. Science 296, 922–927 (2002). References 73 and 74 report that H2A.X loss promotes genome instability and cancer development.
Sheedfar, F. et al. Genetic ablation of macrohistone H2A1 leads to increased leanness, glucose tolerance and energy expenditure in mice fed a high-fat diet. Int. J. Obes. (Lond.) 39, 331–338 (2015).
Boulard, M. et al. Histone variant macroH2A1 deletion in mice causes female-specific steatosis. Epigenetics Chromatin 3, 8 (2010).
Changolkar, L. N. et al. Developmental changes in histone macroH2A1-mediated gene regulation. Mol. Cell. Biol. 27, 2758–2764 (2007).
Pehrson, J. R., Changolkar, L. N., Costanzi, C. & Leu, N. A. Mice without macroH2A histone variants. Mol. Cell. Biol. 34, 4523–4533 (2014).
Creyghton, M. P. et al. H2AZ is enriched at Polycomb complex target genes in ES cells and is necessary for lineage commitment. Cell 135, 649–661 (2008).
Banaszynski, L. A. et al. Hira-dependent histone H3.3 deposition facilitates PRC2 recruitment at developmental loci in ES cells. Cell 155, 107–120 (2013).
Martello, G. & Smith, A. The nature of embryonic stem cells. Annu. Rev. Cell Dev. Biol. 30, 647–675 (2014).
Barrero, M. J., Sese, B., Marti, M. & Izpisua Belmonte, J. C. Macro histone variants are critical for the differentiation of human pluripotent cells. J. Biol. Chem. 288, 16110–16116 (2013).
Hu, G. et al. H2A.Z facilitates access of active and repressive complexes to chromatin in embryonic stem cell self-renewal and differentiation. Cell Stem Cell 12, 180–192 (2013).
Ahuja, A. K. et al. A short G1 phase imposes constitutive replication stress and fork remodelling in mouse embryonic stem cells. Nat. Commun. 7, 10660 (2016).
Turinetto, V. et al. High basal γH2AX levels sustain self-renewal of mouse embryonic and induced pluripotent stem cells. Stem Cells 30, 1414–1423 (2012).
Boskovic, A. et al. Higher chromatin mobility supports totipotency and precedes pluripotency in vivo. Genes Dev. 28, 1042–1047 (2014).
Pasque, V., Jullien, J., Miyamoto, K., Halley-Stott, R. P. & Gurdon, J. B. Epigenetic factors influencing resistance to nuclear reprogramming. Trends Genet. 27, 516–525 (2011).
Jullien, J. et al. HIRA dependent H3.3 deposition is required for transcriptional reprogramming following nuclear transfer to Xenopus oocytes. Epigenetics Chromatin 5, 17 (2012).
Wen, D. et al. Histone variant H3.3 is an essential maternal factor for oocyte reprogramming. Proc. Natl Acad. Sci. USA 111, 7325–7330 (2014).
Ng, R. K. & Gurdon, J. B. Epigenetic memory of an active gene state depends on histone H3.3 incorporation into chromatin in the absence of transcription. Nat. Cell Biol. 10, 102–109 (2008).
Chang, C. C. et al. Rapid elimination of the histone variant macroH2A from somatic cell heterochromatin after nuclear transfer. Cell Reprogram. 12, 43–53 (2010).
Pasque, V., Gillich, A., Garrett, N. & Gurdon, J. B. Histone variant macroH2A confers resistance to nuclear reprogramming. EMBO J. 30, 2373–2387 (2011).
Pasque, V. et al. Histone variant macroH2A marks embryonic differentiation in vivo and acts as an epigenetic barrier to induced pluripotency. J. Cell Sci. 125, 6094–6104 (2012).
Barrero, M. J. et al. Macrohistone variants preserve cell identity by preventing the gain of H3K4me2 during reprogramming to pluripotency. Cell Rep. 3, 1005–1011 (2013).
Gaspar-Maia, A. et al. MacroH2A histone variants act as a barrier upon reprogramming towards pluripotency. Nat. Commun. 4, 1565 (2013). Reference 92 shows that macroH2A contributes to the barrier that inhibits cellular reprogramming mediated by somatic cell nuclear transfer, which was later found — in references 93, 94 and 95 — to also be the case for transcription factor-mediated reprogramming.
Shinagawa, T. et al. Histone variants enriched in oocytes enhance reprogramming to induced pluripotent stem cells. Cell Stem Cell 14, 217–227 (2014).
Zink, L. M. & Hake, S. B. Histone variants: nuclear function and disease. Curr. Opin. Genet. Dev. 37, 82–89 (2016).
Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011).
Cantarino, N., Douet, J. & Buschbeck, M. MacroH2A — an epigenetic regulator of cancer. Cancer Lett. 336, 247–252 (2013).
Kapoor, A. et al. The histone variant macroH2A suppresses melanoma progression through regulation of CDK8. Nature 468, 1105–1109 (2010). This study demonstrates that macroH2A functions as a tumour suppressor in melanoma.
Dardenne, E. et al. Splicing switch of an epigenetic regulator by RNA helicases promotes tumor-cell invasiveness. Nat. Struct. Mol. Biol. 19, 1139–1146 (2012).
Sporn, J. C. et al. Histone macroH2A isoforms predict the risk of lung cancer recurrence. Oncogene 28, 3423–3428 (2009).
Novikov, L. et al. QKI-mediated alternative splicing of the histone variant macroH2A1 regulates cancer cell proliferation. Mol. Cell. Biol. 31, 4244–4255 (2011).
Park, S. J. et al. MacroH2A1 downregulation enhances the stem-like properties of bladder cancer cells by transactivation of lin28B. Oncogene 35, 1292–1301 (2016).
Gallo, M. et al. MLL5 orchestrates a cancer self-renewal state by repressing the histone variant H3.3 and globally reorganizing chromatin. Cancer Cell 28, 715–729 (2015).
Hua, S. et al. Genomic analysis of estrogen cascade reveals histone variant H2A.Z associated with breast cancer progression. Mol. Syst. Biol. 4, 188 (2008).
Yang, H. D. et al. Oncogenic potential of histone-variant H2A.Z.1 and its regulatory role in cell cycle and epithelial–mesenchymal transition in liver cancer. Oncotarget 7, 11412–11423 (2016).
Vardabasso, C. et al. Histone variant H2A.Z.2 mediates proliferation and drug sensitivity of malignant melanoma. Mol. Cell 59, 75–88 (2015). This paper identifies the H2A.Z.2 isoform as a prognostic marker and driver of metastatic melanoma.
Chevillard-Briet, M. et al. Interplay between chromatin-modifying enzymes controls colon cancer progression through Wnt signaling. Hum. Mol. Genet. 23, 2120–2131 (2014).
Muthurajan, U. M., McBryant, S. J., Lu, X., Hansen, J. C. & Luger, K. The linker region of macroH2A promotes self-association of nucleosomal arrays. J. Biol. Chem. 286, 23852–23864 (2011).
Chakravarthy, S., Patel, A. & Bowman, G. D. The basic linker of macroH2A stabilizes DNA at the entry/exit site of the nucleosome. Nucleic Acids Res. 40, 8285–8295 (2012).
Braunschweig, U., Hogan, G. J., Pagie, L. & van Steensel, B. Histone H1 binding is inhibited by histone variant H3.3. EMBO J. 28, 3635–3645 (2009).
White, A. E., Hieb, A. R. & Luger, K. A quantitative investigation of linker histone interactions with nucleosomes and chromatin. Sci. Rep. 6, 19122 (2016).
Izzo, A. & Schneider, R. The role of linker histone H1 modifications in the regulation of gene expression and chromatin dynamics. Biochim. Biophys. Acta 1859, 486–495 (2016).
Kraushaar, D. C. et al. Genome-wide incorporation dynamics reveal distinct categories of turnover for the histone variant H3.3. Genome Biol. 14, R121 (2013).
Gaume, X., Monier, K., Argoul, F., Mongelard, F. & Bouvet, P. In vivo study of the histone chaperone activity of nucleolin by FRAP. Biochem. Res. Int. 2011, 187624 (2011).
Chakravarthy, S. & Luger, K. The histone variant macro-H2A preferentially forms “hybrid nucleosomes”. J. Biol. Chem. 281, 25522–25531 (2006).
Schwartzentruber, J. et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 482, 226–231 (2012).
Wu, G. et al. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat. Genet. 44, 251–253 (2012).
Gessi, M. et al. H3.3 G34R mutations in pediatric primitive neuroectodermal tumors of central nervous system (CNS-PNET) and pediatric glioblastomas: possible diagnostic and therapeutic implications? J. Neurooncol. 112, 67–72 (2013).
Behjati, S. et al. Distinct H3F3A and H3F3B driver mutations define chondroblastoma and giant cell tumor of bone. Nat. Genet. 45, 1479–1482 (2013).
Lewis, P. W. et al. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science 340, 857–861 (2013). This study demonstrates that the histone H3.3 K27M mutation acts through dominant inhibition of PRC2 and suggests the possible existence of analogous mutations of other post-translationally modified residues, which were later found (see reference 128).
Di Croce, L. & Helin, K. Transcriptional regulation by Polycomb group proteins. Nat. Struct. Mol. Biol. 20, 1147–1155 (2013).
Kirmizis, A. et al. Silencing of human Polycomb target genes is associated with methylation of histone H3 Lys 27. Genes Dev. 18, 1592–1605 (2004).
Bender, S. et al. Reduced H3K27me3 and DNA hypomethylation are major drivers of gene expression in K27M mutant pediatric high-grade gliomas. Cancer Cell 24, 660–672 (2013).
Chan, K. M. et al. The histone H3.3K27M mutation in pediatric glioma reprograms H3K27 methylation and gene expression. Genes Dev. 27, 985–990 (2013).
Funato, K., Major, T., Lewis, P. W., Allis, C. D. & Tabar, V. Use of human embryonic stem cells to model pediatric gliomas with H3.3K27M histone mutation. Science 346, 1529–1533 (2014).
Fang, D. et al. The histone H3.3K36M mutation reprograms the epigenome of chondroblastomas. Science 352, 1344–1348 (2016). This study elegantly demonstrates that the K36M mutation of histone H3.3 is a gain-of-function mutation that — analogously to K27M (see reference 122) — inhibits specific histone methyltransferases.
Guo, R. et al. BS69/ZMYND11 reads and connects histone H3.3 lysine 36 trimethylation-decorated chromatin to regulated pre-mRNA processing. Mol. Cell 56, 298–310 (2014).
Rogakou, E. P., Pilch, D. R., Orr, A. H., Ivanova, V. S. & Bonner, W. M. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 273, 5858–5868 (1998). This seminal study identifies the phosphorylation of H2A.X at its unique serine at position 139 (γH2A.X) as a key signal in the DNA damage response.
Bao, Y. Chromatin response to DNA double-strand break damage. Epigenomics 3, 307–321 (2011).
Fernandez-Capetillo, O. et al. DNA damage-induced G2–M checkpoint activation by histone H2AX and 53BP1. Nat. Cell Biol. 4, 993–997 (2002).
Hiom, K. Coping with DNA double strand breaks. DNA Repair (Amst.) 9, 1256–1263 (2010).
Xu, Y. et al. Histone H2A.Z controls a critical chromatin remodeling step required for DNA double-strand break repair. Mol. Cell 48, 723–733 (2012).
Jiang, Y. et al. Local generation of fumarate promotes DNA repair through inhibition of histone H3 demethylation. Nat. Cell Biol. 17, 1158–1168 (2015).
Luijsterburg, M. S. et al. PARP1 links CHD2-mediated chromatin expansion and H3.3 deposition to DNA repair by non-homologous end-joining. Mol. Cell 61, 547–562 (2016).
Khurana, S. et al. A macrohistone variant links dynamic chromatin compaction to BRCA1-dependent genome maintenance. Cell Rep. 8, 1049–1062 (2014).
Mehrotra, P. V. et al. DNA repair factor APLF is a histone chaperone. Mol. Cell 41, 46–55 (2011).
Xu, C., Xu, Y., Gursoy-Yuzugullu, O. & Price, B. D. The histone variant macroH2A1.1 is recruited to DSBs through a mechanism involving PARP1. FEBS Lett. 586, 3920–3925 (2012).
Kusakabe, M. et al. Genetic complementation analysis showed distinct contributions of the N-terminal tail of H2A.Z to epigenetic regulations. Genes Cells 21, 122–135 (2016).
Lovejoy, C. A. et al. Loss of ATRX, genome instability, and an altered DNA damage response are hallmarks of the alternative lengthening of telomeres pathway. PLoS Genet. 8, e1002772 (2012).
Clynes, D. et al. Suppression of the alternative lengthening of telomere pathway by the chromatin remodelling factor ATRX. Nat. Commun. 6, 7538 (2015).
Ramamoorthy, M. & Smith, S. Loss of ATRX suppresses resolution of telomere cohesion to control recombination in ALT cancer cells. Cancer Cell 28, 357–369 (2015).
Creppe, C., Posavec, M., Douet, J. & Buschbeck, M. MacroH2A in stem cells: a story beyond gene repression. Epigenomics 4, 221–227 (2012).
Kim, J. M., Heo, K., Choi, J., Kim, K. & An, W. The histone variant macroH2A regulates Ca2+ influx through TRPC3 and TRPC6 channels. Oncogenesis 2, e77 (2013).
Cong, R. et al. MacroH2A1 histone variant represses rDNA transcription. Nucleic Acids Res. 42, 181–192 (2014).
Chen, H. et al. MacroH2A1 and ATM play opposing roles in paracrine senescence and the senescence-associated secretory phenotype. Mol. Cell 59, 719–731 (2015).
Zilberman, D., Coleman-Derr, D., Ballinger, T. & Henikoff, S. Histone H2A.Z and DNA methylation are mutually antagonistic chromatin marks. Nature 456, 125–129 (2008).
Wolff, E. M. et al. Hypomethylation of a LINE-1 promoter activates an alternate transcript of the MET oncogene in bladders with cancer. PLoS Genet. 6, e1000917 (2010).
Valdes-Mora, F. et al. Acetylation of H2A.Z is a key epigenetic modification associated with gene deregulation and epigenetic remodeling in cancer. Genome Res. 22, 307–321 (2012).
Weyemi, U. et al. The histone variant H2A.X is a regulator of the epithelial–mesenchymal transition. Nat. Commun. 7, 10711 (2016).
Perez-Mancera, P. A., Young, A. R. & Narita, M. Inside and out: the activities of senescence in cancer. Nat. Rev. Cancer 14, 547–558 (2014).
Chan, H. M., Narita, M., Lowe, S. W. & Livingston, D. M. The p400 E1A-associated protein is a novel component of the p53→p21 senescence pathway. Genes Dev. 19, 196–201 (2005).
Gevry, N., Chan, H. M., Laflamme, L., Livingston, D. M. & Gaudreau, L. p21 transcription is regulated by differential localization of histone H2A.Z. Genes Dev. 21, 1869–1881 (2007).
Rai, T. S. et al. HIRA orchestrates a dynamic chromatin landscape in senescence and is required for suppression of neoplasia. Genes Dev. 28, 2712–2725 (2014).
Ye, X. et al. Definition of pRB- and p53-dependent and -independent steps in HIRA/ASF1a-mediated formation of senescence-associated heterochromatin foci. Mol. Cell. Biol. 27, 2452–2465 (2007).
Duarte, L. F. et al. Histone H3.3 and its proteolytically processed form drive a cellular senescence programme. Nat. Commun. 5, 5210 (2014).
Delbarre, E., Ivanauskiene, K., Kuntziger, T. & Collas, P. DAXX-dependent supply of soluble (H3.3–H4) dimers to PML bodies pending deposition into chromatin. Genome Res. 23, 440–451 (2013).
Chang, F. T. et al. PML bodies provide an important platform for the maintenance of telomeric chromatin integrity in embryonic stem cells. Nucleic Acids Res. 41, 4447–4458 (2013).
Korf, K. et al. The PML domain of PML–RARα blocks senescence to promote leukemia. Proc. Natl Acad. Sci. USA 111, 12133–12138 (2014).
Borghesan, M. et al. DNA hypomethylation and histone variant macroH2A1 synergistically attenuate chemotherapy-induced senescence to promote hepatocellular carcinoma progression. Cancer Res. 76, 594–606 (2016).
Zhang, R. et al. Formation of macroH2A-containing senescence-associated heterochromatin foci and senescence driven by ASF1a and HIRA. Dev. Cell 8, 19–30 (2005).
Santoro, S. W. & Dulac, C. The activity-dependent histone variant H2BE modulates the life span of olfactory neurons. eLife 1, e00070 (2012).
Maehara, K. et al. Tissue-specific expression of histone H3 variants diversified after species separation. Epigenetics Chromatin 8, 35 (2015).
Leroy, G. et al. A quantitative atlas of histone modification signatures from human cancer cells. Epigenetics Chromatin 6, 20 (2013).
Hemberger M, Dean W, Reik W. Epigenetic dynamics of stem cells and cell lineage commitment: digging Waddington's canal. Nat. Rev. Mol. Cell Biol. 10, 526–537 (2009).
Belotserkovskaya, R. et al. FACT facilitates transcription-dependent nucleosome alteration. Science 301, 1090–1093 (2003).
Mosammaparast, N., Ewart, C. S. & Pemberton, L. F. A role for nucleosome assembly protein 1 in the nuclear transport of histones H2A and H2B. EMBO J. 21, 6527–6538 (2002).
Andrews, A. J., Chen, X., Zevin, A., Stargell, L. A. & Luger, K. The histone chaperone Nap1 promotes nucleosome assembly by eliminating nonnucleosomal histone DNA interactions. Mol. Cell 37, 834–842 (2010).
Angelov, D. et al. Nucleolin is a histone chaperone with FACT-like activity and assists remodeling of nucleosomes. EMBO J. 25, 1669–1679 (2006).
Alatwi, H. E. & Downs, J. A. Removal of H2A.Z by INO80 promotes homologous recombination. EMBO Rep. 16, 986–994 (2015).
Okuwaki, M., Kato, K., Shimahara, H., Tate, S. & Nagata, K. Assembly and disassembly of nucleosome core particles containing histone variants by human nucleosome assembly protein I. Mol. Cell. Biol. 25, 10639–10651 (2005).
Boulard, M. et al. The NH2 tail of the novel histone variant H2BFWT exhibits properties distinct from conventional H2B with respect to the assembly of mitotic chromosomes. Mol. Cell. Biol. 26, 1518–1526 (2006).
English, C. M., Adkins, M. W., Carson, J. J., Churchill, M. E. & Tyler, J. K. Structural basis for the histone chaperone activity of Asf1. Cell 127, 495–508 (2006).
Tagami, H., Ray-Gallet, D., Almouzni, G. & Nakatani, Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 116, 51–61 (2004).
Elsasser, S. J. et al. DAXX envelops a histone H3.3–H4 dimer for H3.3-specific recognition. Nature 491, 560–565 (2012).
Ray-Gallet, D. et al. HIRA is critical for a nucleosome assembly pathway independent of DNA synthesis. Mol. Cell 9, 1091–1100 (2002).
Ivanauskiene, K. et al. The PML-associated protein DEK regulates the balance of H3.3 loading on chromatin and is important for telomere integrity. Genome Res. 24, 1584–1594 (2014).
Dunleavy, E. M. et al. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell 137, 485–497 (2009).
Foltz, D. R. et al. Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell 137, 472–484 (2009). References 178 and 179 identify and functionally characterize HJURP as CENP-A deposition factor.
Wiedemann, S. M. et al. Identification and characterization of two novel primate-specific histone H3 variants, H3.X and H3.Y. J. Cell Biol. 190, 777–791 (2010).
Postberg, J., Forcob, S., Chang, W. J. & Lipps, H. J. The evolutionary history of histone H3 suggests a deep eukaryotic root of chromatin modifying mechanisms. BMC Evol. Biol. 10, 259 (2010).
Schenk, R., Jenke, A., Zilbauer, M., Wirth, S. & Postberg, J. H3.5 is a novel hominid-specific histone H3 variant that is specifically expressed in the seminiferous tubules of human testes. Chromosoma 120, 275–285 (2011).
Pasque, V., Halley-Stott, R. P., Gillich, A., Garrett, N. & Gurdon, J. B. Epigenetic stability of repressed states involving the histone variant macroH2A revealed by nuclear transfer to Xenopus oocytes. Nucleus 2, 533–539 (2011).
Sansoni, V. et al. The histone variant H2A.Bbd is enriched at sites of DNA synthesis. Nucleic Acids Res. 42, 6405–6420 (2014).
Tolstorukov, M. Y. et al. Histone variant H2A.Bbd is associated with active transcription and mRNA processing in human cells. Mol. Cell 47, 596–607 (2012).
Khuong-Quang, D. A. et al. K27M mutation in histone H3.3 defines clinically and biologically distinct subgroups of pediatric diffuse intrinsic pontine gliomas. Acta Neuropathol. 124, 439–447 (2012).
Forbes, S. A. et al. COSMIC: exploring the world's knowledge of somatic mutations in human cancer. Nucleic Acids Res. 43, D805–D811 (2015).
Acknowledgements
The authors thank E. Bernstein for critical reading of the manuscript and for her insightful suggestions. This work was supported by the Ministry of Economy and Competitiveness (MINECO; grants BFU2015-66559-P and PIE16-00011 to M.B.); the Deutsche Jose Carreras Leukämie Stiftung (grant DJCLS R 14/16 to M.B.); AFM Téléthon (grant AFM 18738 to M.B.); Fundació Internacional Josep Carreras (M.B.); Foundation 'Obra Social la Caixa' (M.B.); Agency for Management of University and Research Grants (AGAUR; grant 2014-SGR-35 to M.B.), the European Commission (grant H2020-MSCA-ITN-2015-675610 to M.B.); the Deutsche Forschungsgemeinschaft (DFG; CRC1064, project A10; grant HA5437/6-1 to S.B.H.); Weigand'sche Stiftung (S.B.H.); and the Center for Integrated Protein Science Munich, Germany (S.B.H.).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Glossary
- Urochordates
-
Also called tunicates. Small marine invertebrates that exhibit a simplified chordate body plan and are within the chordate phylum, which includes the closest relatives of vertebrates.
- Pseudogenes
-
Genes and gene copies that have lost their protein-coding function.
- Histone chaperones
-
Histone-binding proteins that facilitate histone-dependent processes, including histone deposition on and removal from chromatin.
- Chromatin remodellers
-
Frequently multimeric complexes that use ATP to catalyse changes in chromatin structure, including the exchange of histones.
- Retrotransposons
-
Ubiquitous genetic elements that are able to amplify themselves.
- Centromere
-
A chromosome region that mediates attachment to the mitotic spindle during mitosis.
- Inner cell mass
-
Pluripotent cells of the blastocyst.
- Kinetochore
-
A multiprotein complex assembled on the centromere that mediates the interaction with the microtubules of the spindle.
- Totipotent cells
-
Cells that can give rise to all embryonic and extra-embryonic tissues.
- Epithelial–mesenchymal transition
-
(EMT). The switch of cells from an epithelial to a mesenchymal morphology.
- G2–M checkpoint
-
A control mechanism that allows cell cycle progression only in the absence of DNA damage.
- Homologous recombination
-
The exchange of DNA sequences between different chromosome copies.
- Non-homologous end joining
-
(NHEJ). A DNA repair pathway that ligates DNA break ends independently of their sequence.
- Neocentromeres
-
New centromeres that form on chromosome arms.
- Pericentromeric heterochromatin
-
Heterochromatin regions that flank the centromere.
- Alternative lengthening of telomeres pathway
-
(ALT pathway). A telomerase-independent mechanism of telomere maintenance in proliferating cells.
- G quadruplexes
-
Stable non-helical tertiary structures formed from guanine-rich nucleic acid sequences.
- rDNA clusters
-
Arrays of repeated sequences that encode the RNA components of ribosomes.
- Paracrine senescence
-
Irreversible cell cycle arrest induced by paracrine factors that are secreted by other senescent cells.
- Long interspersed element 1 repeat elements
-
(LINE1 repeat elements). A large family of mammalian retrotransposons.
- Oncogene-induced senescence
-
Irreversible cell cycle arrest induced by acute oncogene overexpression.
- Promyelocytic leukaemia bodies
-
(PML bodies). Nuclear bodies formed by the protein PML and multiple other proteins that interact with PML.
Rights and permissions
About this article
Cite this article
Buschbeck, M., Hake, S. Variants of core histones and their roles in cell fate decisions, development and cancer. Nat Rev Mol Cell Biol 18, 299–314 (2017). https://doi.org/10.1038/nrm.2016.166
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrm.2016.166
This article is cited by
-
The esBAF and ISWI nucleosome remodeling complexes influence occupancy of overlapping dinucleosomes and fragile nucleosomes in murine embryonic stem cells
BMC Genomics (2023)
-
Chromatin balances cell redox and energy homeostasis
Epigenetics & Chromatin (2023)
-
(B)On(e)-cohistones and the epigenetic alterations at the root of bone cancer
Cell Death & Differentiation (2023)
-
Epigenetic control of cancer inflammation
Nature Cell Biology (2023)
-
Deficiency of histone variant macroH2A1.1 is associated with sexually dimorphic obesity in mice
Scientific Reports (2023)