Abstract
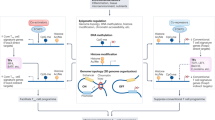
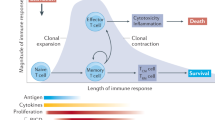
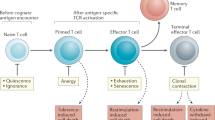
Regulatory T (TReg) cells are crucial for the prevention of fatal autoimmunity in mice and humans. Forkhead box P3 (FOXP3)+ TReg cells are produced in the thymus and are also generated from conventional CD4+ T cells in peripheral sites. It has been suggested that FOXP3+ TReg cells might become unstable under certain inflammatory conditions and might adopt a phenotype that is more characteristic of effector CD4+ T cells. These suggestions have caused considerable debate in the field and have important implications for the therapeutic use of TReg cells. In this article, Nature Reviews Immunology asks several experts for their views on the plasticity and stability of TReg cells.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Sakaguchi, S., Yamaguchi, T., Nomura, T. & Ono, M. Regulatory T cells and immune tolerance. Cell 133, 775–787 (2008).
Duarte, J. H., Zelenay, S., Bergman, M. L., Martins, A. C. & Demengeot, J. Natural Treg cells spontaneously differentiate into pathogenic helper cells in lymphopenic conditions. Eur. J. Immunol. 39, 948–955 (2009).
Miyao, T. et al. Plasticity of Foxp3+ T cells reflects promiscuous Foxp3 expression in conventional T cells but not reprogramming of regulatory T cells. Immunity. 36, 262–275 (2012).
Vignali, D. A. A., Collison, L. W. & Workman, C. J. How regulatory T cells work. Nature Rev. Immunol. 8, 523–532 (2008).
Benoist, C. & Mathis, D. Treg cells, life history, and diversity. Cold Spring Harb. Perspect. Biol. 4, a007021 (2012).
Rudensky, A. Y. Regulatory T cells and Foxp3. Immunol. Rev. 241, 260–268 (2011).
Fu, W. et al. A multiply redundant genetic switch 'locks in' the transcriptional signature of regulatory T cells. Nature Immunol. 13, 972–980 (2012).
Ohkura, N. et al. T cell receptor stimulation-induced epigenetic changes and Foxp3 expression are independent and complementary events required for Treg cell development. Immunity 37, 785–799 (2012).
Sakaguchi, S. Regulatory T cells: history and perspective. Methods Mol. Biol. 707, 3–17 (2011).
Workman, C. J., Szymczak-Workman, A. L., Collison, L. W., Pillai, M. R. & Vignali, D. A. The development and function of regulatory T cells. Cell. Mol. Life Sci. 66, 2603–2622 (2009).
Collison, L. W., Pillai, M. R., Chaturvedi, V. & Vignali, D. A. Regulatory T cell suppression is potentiated by target T cells in a cell contact, IL-35- and IL-10-dependent manner. J. Immunol. 182, 6121–6128 (2009).
Abbas, A. K. et al. Regulatory T cells: recommendations to simplify the nomenclature. Nature Immunol. 14, 307–308 (2013).
Rubtsov, Y. P. et al. Stability of the regulatory T cell lineage in vivo. Science 329, 1667–1671 (2010).
Zhou, X. et al. Instability of the transcription factor Foxp3 leads to the generation of pathogenic memory T cells in vivo. Nature Immunol. 10, 1000–1007 (2009).
Hori, S., Nomura, T. & Sakaguchi, S. Control of regulatory T cell development by the transcription factor Foxp3. Science 299, 1057–1061 (2003).
Fontenot, J. D. et al. Regulatory T cell lineage specification by the forkhead transcription factor foxp3. Immunity 22, 329–341 (2005).
Williams, L. M. & Rudensky, A. Y. Maintenance of the Foxp3-dependent developmental program in mature regulatory T cells requires continued expression of Foxp3. Nature Immunol. 8, 277–284 (2007).
Samstein, R. M. et al. Foxp3 exploits a pre-existent enhancer landscape for regulatory T cell lineage specification. Cell 151, 153–166 (2012).
Hori, S. Stability of regulatory T-cell lineage. Adv. Immunol. 112, 1–24 (2011).
Sakaguchi, S., Miyara, M., Costantino, C. M. & Hafler, D. A. FOXP3+ regulatory T cells in the human immune system. Nature Rev. Immunol. 10, 490–500 (2010).
Tian, L., Humblet-Baron, S. & Liston, A. Immune tolerance: are regulatory T cell subsets needed to explain suppression of autoimmunity? BioEssays 34, 569–575 (2012).
Nie, H. et al. Phosphorylation of FOXP3 controls regulatory T cell function and is inhibited by TNF-α in rheumatoid arthritis. Nature Med. 19, 322–328 (2013).
Zhou, X., Bailey-Bucktrout, S., Jeker, L. T. & Bluestone, J. A. Plasticity of CD4+ FoxP3+ T cells. Curr. Opin. Immunol. 21, 281–285 (2009).
Lal, G. et al. Distinct inflammatory signals have physiologically divergent effects on epigenetic regulation of Foxp3 expression and Treg function. Am. J. Transplant. 11, 203–214 (2011).
Yurchenko, E. et al. Inflammation-driven reprogramming of CD4+ Foxp3+ regulatory T cells into pathogenic Th1/Th17 T effectors is abrogated by mTOR inhibition in vivo. PLoS ONE 7, e35572 (2012).
d'Hennezel, E., Yurchenko, E., Sgouroudis, E., Hay, V. & Piccirillo, C. A. Single-cell analysis of the human T regulatory population uncovers functional heterogeneity and instability within FOXP3+ cells. J. Immunol. 186, 6788–6797 (2011).
Pasare, C. & Medzhitov, R. Toll pathway-dependent blockade of CD4+CD25+ T cell-mediated suppression by dendritic cells. Science 299, 1033–1036 (2003).
Koch, M. A. et al. The transcription factor T-bet controls regulatory T cell homeostasis and function during type 1 inflammation. Nature Immunol. 10, 595–602 (2009).
Chaudhry, A. et al. CD4+ regulatory T cells control TH17 responses in a Stat3-dependent manner. Science 326, 986–991 (2009).
Zheng, Y. et al. Regulatory T-cell suppressor program co-opts transcription factor IRF4 to control TH2 responses. Nature 458, 351–356 (2009).
Polansky, J. K. et al. DNA methylation controls Foxp3 gene expression. Eur. J. Immunol. 38, 1654–1663 (2008).
Zheng, Y. et al. Role of conserved non-coding DNA elements in the Foxp3 gene in regulatory T-cell fate. Nature 463, 808–812 (2010).
Josefowicz, S. Z. et al. Extrathymically generated regulatory T cells control mucosal TH2 inflammation. Nature 482, 395–399 (2012).
Josefowicz, S. Z., Lu, L.-F. & Rudensky, A. Y. Regulatory T cells: mechanisms of differentiation and function. Annu. Rev. Immunol. 30, 531–564 (2012).
Oldenhove, G. et al. Decrease of Foxp3+ Treg cell number and acquisition of effector cell phenotype during lethal infection. Immunity 31, 772–786 (2009).
Floess, S. et al. Epigenetic control of the foxp3 locus in regulatory T cells. PLoS Biol. 5, e38 (2007).
Wei, G. et al. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity 30, 155–167 (2009).
Takahashi, T. et al. Immunologic self-tolerance maintained by CD25+CD4+ naturally anergic and suppressive T cells: induction of autoimmune disease by breaking their anergic/suppressive state. Int. Immunol. 10, 1969–1980 (1998).
Thornton, A. M. & Shevach, E. M. CD4+CD25+ immunoregulatory T cells suppress polyclonal T cell activation in vitro by inhibiting interleukin 2 production. J. Exp. Med. 188, 287–296 (1998).
Daniel, C., Wennhold, K., Kim, H. J. & von Boehmer, H. Enhancement of antigen-specific Treg vaccination in vivo. Proc. Natl Acad. Sci. USA 107, 16246–16251 (2010).
Cobbold, S. P., Adams, E., Marshall, S. E., Davies, J. D. & Waldmann, H. Mechanisms of peripheral tolerance and suppression induced by monoclonal antibodies to CD4 and CD8. Immunol. Rev. 149, 5–33 (1996).
Chang, J. T. et al. Asymmetric T lymphocyte division in the initiation of adaptive immune responses. Science 315, 1687–1691 (2007).
Miyara, M. et al. Functional delineation and differentiation dynamics of human CD4+ T cells expressing the FoxP3 transcription factor. Immunity 30, 899–911 (2009).
Kendal, A. R. et al. Sustained suppression by Foxp3+ regulatory T cells is vital for infectious transplantation tolerance. J. Exp. Med. 208, 2043–2053 (2011).
Beier, U. H. et al. Histone deacetylases 6 and 9 and sirtuin-1 control Foxp3+ regulatory T cell function through shared and isoform-specific mechanisms. Sci. Signal. 5, ra45 (2012).
Zhang, H., Xiao, Y., Zhu, Z., Li, B. & Greene, M. I. Immune regulation by histone deacetylases: a focus on the alteration of FOXP3 activity. Immunol. Cell Biol. 90, 95–100 (2012).
Wing, J. B. & Sakaguchi, S. Multiple Treg suppressive modules and their adaptability. Front. Immunol. 3, 178 (2012).
Wang, H. Y. & Wang, R. F. Regulatory T cells and cancer. Curr. Opin. Immunol. 19, 217–223 (2007).
Acknowledgements
S.S. acknowledges the daily discussions he had with his colleagues that helped him to write his comments for this article.
D.A.A.V. apologises to those investigators who contributed important observations related to the questions he covered that he could not discuss or quote owing to space limitations. D.A.A.V. is supported by the US National Institutes of Health (NIH) (grants AI091977, AI039480, AI052199, DK089125), American Asthma Foundation (grant10-0128), National Cancer Institute Comprehensive Cancer Center grant (CA21765) and ALSAC.
A.Y.R. is supported by an NIH grant (R37 AI034206) and is an investigator at the Howard Hughes Medical Center. R.E.N. is supported by an NIH Medical Scientist Training Program grant (GM07739) and a National Institute of Neurological Disorders and Stroke grant (1F31NS073203-01).
H.W. wishes to acknowledge the help of his colleagues D. Howie, R. Hilbrands and S. Cobbold in writing his comments for this article.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
D.A.A.V. declares competing financial interests. He has submitted patents that are pending or granted, and is entitled to a share in net income generated from licensing of these patent rights for commercial development. He also consults for several biopharmaceutical companies.
S.S., R.E.N., A.R. and H.W. declare no competing financial interests.
Related links
Glossary
- Asymmetric T cell division
-
A process by which two daughter cells can inherit different amounts of immune receptors and signalling components from a parent cell during T cell division. It has been suggested that this process occurs because of the polarity of the dividing cell that is associated with immunological synapse formation and that it could specify different fates to the progeny of an individual T cell.
- Conserved non-coding sequence 2
-
(CNS2). The element engaged in the positive auto-feedback loop that confers heritable maintenance of forkhead box P3 (FOXP3) expression once demethylated. Also known as the TSDR (TReg cell-specific demethylated region).
- CpG island
-
A sequence of 0.5–2 kilobases that is rich in CpG dinucleotides. CpG islands are mostly located upstream of housekeeping genes and also upstream of some tissue-specific genes. They are constitutively non-methylated in all animal cell types.
- Infectious tolerance
-
The ability of a tolerized population of T cells to induce tolerance in a new, naive population of T cells. Tolerance might be to the same antigens or to new antigens that are encountered in the same context (linked suppression). Newly tolerized T cells can, in turn, induce tolerance in other T cells.
- NOD mice
-
Non-obese diabetic (NOD) mice spontaneously develop a form of autoimmunity that closely resembles human type 1 diabetes.
Rights and permissions
About this article
Cite this article
Sakaguchi, S., Vignali, D., Rudensky, A. et al. The plasticity and stability of regulatory T cells. Nat Rev Immunol 13, 461–467 (2013). https://doi.org/10.1038/nri3464
Published:
Issue Date:
DOI: https://doi.org/10.1038/nri3464
This article is cited by
-
The role of transcription factors in shaping regulatory T cell identity
Nature Reviews Immunology (2023)
-
Tumor microenvironment antigens
Seminars in Immunopathology (2023)
-
Oxidative stress and neuroimmune proteins in a mouse model of autism
Cell Stress and Chaperones (2023)
-
Immune Cells in Cardiac Injury Repair and Remodeling
Current Cardiology Reports (2023)
-
FOXP3 expression in esophageal squamous cell carcinoma
Wiener klinische Wochenschrift (2023)