Key Points
-
The macrophage lineage includes a remarkable variety of cells with different functions and functional states that are specified by the complex interplay between microenvironmental signals and a hardwired differentiation programme that determines macrophage identity.
-
The global regulatory landscape of macrophage-specific genes is controlled by the ETS family transcription factor PU.1, which dictates the general context in which sequence-specific transcription factors that are regulated by external stimuli operate to modulate macrophage phenotype and function.
-
Different sequence-specific transcription factors regulate macrophage polarization in specific contexts. Signal transducer and activator of transcription (STAT) proteins are pivotal factors in M1 and M2 macrophage polarization in response to T cell-derived cytokines; peroxisome proliferator-activated receptor-γ regulates the M2-like phenotype of macrophages in adipose tissue; and CCAAT/enhancer-binding protein-β activation mediated by cAMP-responsive element-binding protein drives the induction of M2 macrophages in muscle injury.
-
The polarized phenotype of macrophages is reinforced by the mutual exclusivity of signalling pathways and reciprocal regulation of M1 and M2 genes.
-
Tumour-associated macrophages (TAMs) are a unique polarized macrophage population characterized by the expression of both M1 and M2 marker genes. The specific transcriptional mechanisms that regulate the TAM phenotype remain poorly defined.
Abstract
In terms of both phenotype and function, macrophages have remarkable heterogeneity, which reflects the specialization of tissue-resident macrophages in microenvironments as different as liver, brain and bone. Also, marked changes in the activity and gene expression programmes of macrophages can occur when they come into contact with invading microorganisms or injured tissues. Therefore, the macrophage lineage includes a remarkable diversity of cells with different functions and functional states that are specified by a complex interplay between microenvironmental signals and a hardwired differentiation programme that determines macrophage identity. In this Review, we summarize the current knowledge of transcriptional and chromatin-mediated control of macrophage polarization in physiology and disease.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Auffray, C., Sieweke, M. H. & Geissmann, F. Blood monocytes: development, heterogeneity, and relationship with dendritic cells. Annu. Rev. Immunol. 27, 669–692 (2009).
Gordon, S. & Taylor, P. R. Monocyte and macrophage heterogeneity. Nature Rev. Immunol. 5, 953–964 (2005).
Cheong, C. et al. Microbial stimulation fully differentiates monocytes to DC-SIGN/CD209+ dendritic cells for immune T cell areas. Cell 143, 416–429 (2010). This paper established that monocyte-derived DCs, rather than non-myeloid DCs, are required for CD4+ and CD8+ T cell priming in the context of inflammation.
Sallusto, F. & Lanzavecchia, A. Monocytes join the dendritic cell family. Cell 143, 339–340 (2010).
Gordon, S. The macrophage: past, present and future. Eur. J. Immunol. 37, S9–S17 (2007).
Mosser, D. M. & Edwards, J. P. Exploring the full spectrum of macrophage activation. Nature Rev. Immunol. 8, 958–969 (2008).
Toshchakov, V. et al. TLR4, but not TLR2, mediates IFN-β-induced STAT1α/β-dependent gene expression in macrophages. Nature Immunol. 3, 392–398 (2002).
Condeelis, J. & Pollard, J. W. Macrophages: obligate partners for tumor cell migration, invasion, and metastasis. Cell 124, 263–266 (2006).
Martinez, F. O., Gordon, S., Locati, M. & Mantovani, A. Transcriptional profiling of the human monocyte-to-macrophage differentiation and polarization: new molecules and patterns of gene expression. J. Immunol. 177, 7303–7311 (2006). This study is one of only a few to look at the transcriptome of human polarized macrophage subsets.
Biswas, S. K. et al. A distinct and unique transcriptional program expressed by tumor-associated macrophages (defective NF-κB and enhanced IRF-3/STAT1 activation). Blood 107, 2112–2122 (2006). This was the first description of the specific transcriptional profile of TAMs.
Foster, S. L., Hargreaves, D. C. & Medzhitov, R. Gene-specific control of inflammation by TLR-induced chromatin modifications. Nature 447, 972–978 (2007).
Biswas, S. K. & Lopez-Collazo, E. Endotoxin tolerance: new mechanisms, molecules and clinical significance. Trends Immunol. 30, 475–487 (2009).
Rosenbauer, F. & Tenen, D. G. Transcription factors in myeloid development: balancing differentiation with transformation. Nature Rev. Immunol. 7, 105–117 (2007).
Olson, M. C. et al. PU.1 is not essential for early myeloid gene expression but is required for terminal myeloid differentiation. Immunity 3, 703–714 (1995).
Nerlov, C. & Graf, T. PU.1 induces myeloid lineage commitment in multipotent hematopoietic progenitors. Genes Dev. 12, 2403–2412 (1998). References 14 and 15 demonstrated the essential role of PU.1 in the control of macrophage differentiation.
DeKoter, R. P. & Singh, H. Regulation of B lymphocyte and macrophage development by graded expression of PU.1. Science 288, 1439–1441 (2000).
Chen, X. et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133, 1106–1117 (2008).
Tijssen, M. R. et al. Genome-wide analysis of simultaneous GATA1/2, RUNX1, FLI1, and SCL binding in megakaryocytes identifies hematopoietic regulators. Dev. Cell 20, 597–609 (2011).
Siersbaek, R. et al. Extensive chromatin remodelling and establishment of transcription factor 'hotspots' during early adipogenesis. EMBO J. 30, 1459–1472 (2011).
Heintzman, N. et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nature Genet. 39, 311–318 (2007). This was the first paper to show the distinctive chromatin features of enhancers using high-throughput approaches.
Heintzman, N. D. et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459, 108–112 (2009).
Creyghton, M. P. et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl Acad. Sci. USA 107, 21931–21936 (2010).
Rada-Iglesias, A. et al. A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470, 279–283 (2011).
Boyle, A. P. et al. High-resolution mapping and characterization of open chromatin across the genome. Cell 132, 311–322 (2008).
Ghisletti, S. et al. Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity 32, 317–328 (2010).
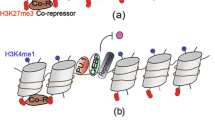
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010). References 25 and 26 report the first characterization of enhancers in macrophages, identifying PU.1 as a main player.
Natoli, G. Maintaining cell identity through global control of genomic organization. Immunity 33, 12–24 (2010).
Holtschke, T. et al. Immunodeficiency and chronic myelogenous leukemia-like syndrome in mice with a targeted mutation of the ICSBP gene. Cell 87, 307–317 (1996).
Krausgruber, T. et al. IRF5 promotes inflammatory macrophage polarization and TH1–TH17 responses. Nature Immunol. 12, 231–238 (2011). This important study first described the requirement for IRF5 in the control of M1 macrophage polarization.
Satoh, T. et al. The Jmjd3–Irf4 axis regulates M2 macrophage polarization and host responses against helminth infection. Nature Immunol. 11, 936–944 (2010). This study demonstrated the role of JMJD3-mediated IRF4 activation in driving M2 macrophage polarization in response to helminths.
Ruffell, D. et al. A CREB–C/EBPβ cascade induces M2 macrophage-specific gene expression and promotes muscle injury repair. Proc. Natl Acad. Sci. USA 106, 17475–17480 (2009). This elegant study showed that the CREB-dependent induction of C/EBPβ expression is specifically required for M2-like macrophage polarization.
De Santa, F. et al. Jmjd3 contributes to the control of gene expression in LPS-activated macrophages. EMBO J. 28, 3341–3352 (2009).
Ramirez-Carrozzi, V. R. et al. A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling. Cell 138, 114–128 (2009).
Hargreaves, D. C., Horng, T. & Medzhitov, R. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell 138, 129–145 (2009). References 33 and 34 demonstrated that the presence of a CpG island relieves dependence on chromatin remodelling for inducible gene activation.
Deaton, A. M. & Bird, A. CpG islands and the regulation of transcription. Genes Dev. 25, 1010–1022 (2009).
Ayton, P. M., Chen, E. H. & Cleary, M. L. Binding to nonmethylated CpG DNA is essential for target recognition, transactivation, and myeloid transformation by an MLL oncoprotein. Mol. Cell. Biol. 24, 10470–10478 (2004).
Lee, J. H. & Skalnik, D. G. CpG-binding protein (CXXC finger protein 1) is a component of the mammalian Set1 histone H3-Lys4 methyltransferase complex, the analogue of the yeast Set1/COMPASS complex. J. Biol. Chem. 280, 41725–41731 (2005).
Lee, J. H. & Skalnik, D. G. Wdr82 is a C-terminal domain-binding protein that recruits the Setd1A histone H3-Lys4 methyltransferase complex to transcription start sites of transcribed human genes. Mol. Cell. Biol. 28, 609–618 (2008).
Valouev, A. et al. Determinants of nucleosome organization in primary human cells. Nature 474, 516–520 (2011).
Ramirez-Carrozzi, V. R. et al. Selective and antagonistic functions of SWI/SNF and Mi-2β nucleosome remodeling complexes during an inflammatory response. Genes Dev. 20, 282–296 (2006). This was the first description of a required role for SWI/SNF-dependent chromatin remodelling in pro-inflammatory gene induction.
Kayama, H. et al. Class-specific regulation of pro-inflammatory genes by MyD88 pathways and IκBζ. J. Biol. Chem. 283, 12468–12477 (2008).
Nau, G. J. et al. Human macrophage activation programs induced by bacterial pathogens. Proc. Natl Acad. Sci. USA 99, 1503–1508 (2002).
Nicodeme, E. et al. Suppression of inflammation by a synthetic histone mimic. Nature 468, 1119–1123 (2011). This report first demonstrated the possibility of selectively interfering with a subset of pro-inflammatory genes by exploiting chromatin-dependent regulation.
Smale, S. T. Selective transcription in response to an inflammatory stimulus. Cell 140, 833–844 (2010).
Darnell, J. E., Jr, Kerr, I. M. & Stark, G. R. Jak–STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 264, 1415–1421 (1994).
Park, C., Li, S., Cha, E. & Schindler, C. Immune response in Stat2 knockout mice. Immunity 13, 795–804 (2000).
Meraz, M. A. et al. Targeted disruption of the Stat1 gene in mice reveals unexpected physiologic specificity in the JAK–STAT signaling pathway. Cell 84, 431–442 (1996).
Durbin, J. E., Hackenmiller, R., Simon, M. C. & Levy, D. E. Targeted disruption of the mouse Stat1 gene results in compromised innate immunity to viral disease. Cell 84, 443–450 (1996).
Varinou, L. et al. Phosphorylation of the Stat1 transactivation domain is required for full-fledged IFN-γ-dependent innate immunity. Immunity 19, 793–802 (2003).
Kovarik, P., Sauer, I. & Schaljo, B. Molecular mechanisms of the anti-inflammatory functions of interferons. Immunobiology 212, 895–901 (2007).
Stockinger, S. et al. Characterization of the interferon-producing cell in mice infected with Listeria monocytogenes. PLoS Pathog. 5, e1000355 (2009).
Kaplan, D. H. et al. Demonstration of an interferon γ-dependent tumor surveillance system in immunocompetent mice. Proc. Natl Acad. Sci. USA 95, 7556–7561 (1998).
Martinez, F. O., Helming, L. & Gordon, S. Alternative activation of macrophages: an immunologic functional perspective. Annu. Rev. Immunol. 27, 451–483 (2009).
Herbert, D. R. et al. Alternative macrophage activation is essential for survival during schistosomiasis and downmodulates T helper 1 responses and immunopathology. Immunity 20, 623–635 (2004).
Brombacher, F., Arendse, B., Peterson, R., Holscher, A. & Holscher, C. Analyzing classical and alternative macrophage activation in macrophage/neutrophil-specific IL-4 receptor-α-deficient mice. Methods Mol. Biol. 531, 225–252 (2009).
Takeda, K. et al. Essential role of Stat6 in IL-4 signalling. Nature 380, 627–630 (1996).
Ohmori, Y. & Hamilton, T. A. IL-4-induced STAT6 suppresses IFN-γ-stimulated STAT1-dependent transcription in mouse macrophages. J. Immunol. 159, 5474–5482 (1997).
Fruman, D. A. et al. Impaired B cell development and proliferation in absence of phosphoinositide 3-kinase p85α. Science 283, 393–397 (1999).
Rauh, M. J. et al. SHIP represses the generation of alternatively activated macrophages. Immunity 23, 361–374 (2005).
DeNardo, D. G. et al. CD4+ T cells regulate pulmonary metastasis of mammary carcinomas by enhancing protumor properties of macrophages. Cancer Cell 16, 91–102 (2009).
Gordon, S. & Martinez, F. O. Alternative activation of macrophages: mechanism and functions. Immunity 32, 593–604 (2010).
Olefsky, J. M. & Glass, C. K. Macrophages, inflammation, and insulin resistance. Annu. Rev. Physiol. 72, 219–246 (2010).
Ricote, M., Li, A. C., Willson, T. M., Kelly, C. J. & Glass, C. K. The peroxisome proliferator-activated receptor-γ is a negative regulator of macrophage activation. Nature 391, 79–82 (1998). This was the first description of the anti-inflammatory function of PPARγ through transrepression of NF-κB and AP1.
Pascual, G. et al. A SUMOylation-dependent pathway mediates transrepression of inflammatory response genes by PPAR-γ. Nature 437, 759–763 (2005).
Bouhlel, M. A. et al. PPARγ activation primes human monocytes into alternative M2 macrophages with anti-inflammatory properties. Cell Metab. 6, 137–143 (2007).
Odegaard, J. I. et al. Macrophage-specific PPARγ controls alternative activation and improves insulin resistance. Nature 447, 1116–1120 (2007). References 65 and 66 demonstrated the requirement for PPARγ in M2 macrophage polarization.
Huang, J. T. et al. Interleukin-4-dependent production of PPAR-γ ligands in macrophages by 12/15-lipoxygenase. Nature 400, 378–382 (1999).
Szanto, A. et al. STAT6 transcription factor is a facilitator of the nuclear receptor PPARγ-regulated gene expression in macrophages and dendritic cells. Immunity 33, 699–712 (2010). This study demonstrated that two key players in M2 macrophage polarization — STAT6 and PPARγ — functionally and physically interact.
Van Ginderachter, J. A. et al. Peroxisome proliferator-activated receptor γ (PPARγ) ligands reverse CTL suppression by alternatively activated (M2) macrophages in cancer. Blood 108, 525–535 (2006).
Friedman, A. D. Transcriptional control of granulocyte and monocyte development. Oncogene 26, 6816–6828 (2007).
El Kasmi, K. C. et al. Toll-like receptor-induced arginase 1 in macrophages thwarts effective immunity against intracellular pathogens. Nature Immunol. 9, 1399–1406 (2008).
Hu, H. M., Baer, M., Williams, S. C., Johnson, P. F. & Schwartz, R. C. Redundancy of C/EBPα, -β, and -δ in supporting the lipopolysaccharide-induced transcription of IL-6 and monocyte chemoattractant protein-1. J. Immunol. 160, 2334–2342 (1998).
Gorgoni, B., Maritano, D., Marthyn, P., Righi, M. & Poli, V. C/EBPβ gene inactivation causes both impaired and enhanced gene expression and inverse regulation of IL-12 p40 and p35 mRNAs in macrophages. J. Immunol. 168, 4055–4062 (2002).
Kim, C. et al. Antiinflammatory cAMP signaling and cell migration genes co-opted by the anthrax bacillus. Proc. Natl Acad. Sci. USA 105, 6150–6155 (2008).
Ananieva, O. et al. The kinases MSK1 and MSK2 act as negative regulators of Toll-like receptor signaling. Nature Immunol. 9, 1028–1036 (2008).
Marigo, I. et al. Tumor-induced tolerance and immune suppression depend on the C/EBPβ transcription factor. Immunity 32, 790–802 (2010). An important study demonstrating that MDSC production and activity are crucially linked to C/EBPβ function.
Youn, J. I. & Gabrilovich, D. I. The biology of myeloid-derived suppressor cells: the blessing and the curse of morphological and functional heterogeneity. Eur. J. Immunol. 40, 2969–2975 (2010).
Gallina, G. et al. Tumors induce a subset of inflammatory monocytes with immunosuppressive activity on CD8+ T cells. J. Clin. Invest. 116, 2777–2790 (2006). The first description of MDSCs in the context of tumour-induced immune suppression.
Savitsky, D., Tamura, T., Yanai, H. & Taniguchi, T. Regulation of immunity and oncogenesis by the IRF transcription factor family. Cancer Immunol. Immunother. 59, 489–510 (2010).
De Santa, F. et al. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell 130, 1083–1094 (2007). This study reports the cloning of Jmjd3 and provides an initial description of its role in macrophage function.
Ishii, M. et al. Epigenetic regulation of the alternatively activated macrophage phenotype. Blood 114, 3244–3254 (2009).
El Chartouni, C., Schwarzfischer, L. & Rehli, M. Interleukin-4 induced interferon regulatory factor (Irf) 4 participates in the regulation of alternative macrophage priming. Immunobiology 215, 821–825 (2010).
Takaoka, A. et al. Integral role of IRF-5 in the gene induction programme activated by Toll-like receptors. Nature 434, 243–249 (2005).
Fleetwood, A. J., Lawrence, T., Hamilton, J. A. & Cook, A. D. Granulocyte-macrophage colony-stimulating factor (CSF) and macrophage CSF-dependent macrophage phenotypes display differences in cytokine profiles and transcription factor activities: implications for CSF blockade in inflammation. J. Immunol. 178, 5245–5252 (2007).
Gilchrist, M. et al. Systems biology approaches identify ATF3 as a negative regulator of Toll-like receptor 4. Nature 441, 173–178 (2006).
Geissmann, F., Jung, S. & Littman, D. R. Blood monocytes consist of two principal subsets with distinct migratory properties. Immunity 19, 71–82 (2003). This was the first characterization of two distinct populations of blood monocytes with different recruitment and functional properties.
Nahrendorf, M. et al. The healing myocardium sequentially mobilizes two monocyte subsets with divergent and complementary functions. J. Exp. Med. 204, 3037–3047 (2007).
Arnold, L. et al. Inflammatory monocytes recruited after skeletal muscle injury switch into antiinflammatory macrophages to support myogenesis. J. Exp. Med. 204, 1057–1069 (2007). An excellent illustration of the dynamic nature of mononuclear phagocyte phenotype during inflammation.
Acknowledgements
T.L. is a research director of the Institut National de la Santé et de la Recherche Médicale (INSERM) supported by grants from the European Research Council (ERC) and the Agence Nationale de la Recherche (ANR), and institutional grants from INSERM, the Centre National de la Recherche Scientifique (CNRS) and the Université de la Méditerranée. Research in this area in the laboratory of G.N. is supported by the ERC.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Glossary
- Mononuclear phagocyte system
-
(MPS). A group of bone marrow-derived cells with different morphologies (monocytes, macrophages and dendritic cells), which are mainly responsible for phagocytosis, cytokine secretion and antigen presentation.
- M1 macrophages
-
Macrophages with an activation phenotype associated with increased microbicidal activity and antigen-presenting function. M1-type activation is usually modelled in vitro by interferon-γ and/or lipopolysaccharide stimulation. In mice, M1-associated markers include interleukin-12, MHC class II molecules and nitric oxide synthase 2 (NOS2); however, human macrophages do not show induction of NOS2 in these conditions, despite having a similar functional phenotype.
- M2 macrophages
-
Macrophages that are associated with parasitic infections and T helper 2-type immune responses. M2-type activation is usually modelled in vitro by interleukin-4 (IL-4) and/or IL-13 stimulation. In mice, M2-associated markers include resistin-like-α (also known as FIZZ1), arginase 1, chitinase 3-like 3 (also known as YM1), IL-10 and macrophage mannose receptor 1 (also known as CD206); however, human M2 macrophages do not show induction of resistin-like-α, arginase 1 and chitinase 3-like 3, but instead upregulate indoleamine 2,3-dioxygenase expression. M2 macrophages are associated with anti-inflammatory and homeostatic functions linked to wound healing, fibrosis and tissue repair.
- Chromatin immunoprecipitation
-
(ChIP). A powerful method for assessing the physical association of a known nuclear protein with a candidate target locus in vivo. Cells are first treated with an agent that crosslinks protein to DNA. The chromatin is then sheared into fragments and the protein is immunoprecipitated. If the candidate target region is co-precipitated (as measured by PCR), the target locus is likely to bind the protein (directly or indirectly) in vivo. The immunoprecipitated DNA can also be used for hybridization to high-density microarrays (ChIP–chip) or for high-throughput sequencing (ChIP–seq) to provide a genome-wide view of the binding sites of a specific transcriptional regulator.
- Gene promoters
-
The regulatory regions to which RNA polymerase binds to initiate transcription. Upstream of the RNA polymerase recruitment site (core promoter) is a regulatory promoter where transcription factors bind to control recruitment of the transcriptional machinery.
- Enhancers
-
Control elements located at variable distances from the genes they regulate and to which multiple regulatory proteins bind, thereby influencing gene transcription. In vitro, enhancers function in an orientation- and position-independent manner, but it is not clear whether this is also true in vivo.
- Primary response genes
-
Inducible genes whose transcription does not require new protein synthesis.
- Secondary response genes
-
Inducible genes whose transcription requires new protein synthesis (often the synthesis of transcription factors required for the activation of these genes).
- SWI/SNF complex
-
An ATP-dependent chromatin-remodelling protein complex that was initially identified in yeast. Related complexes exist in mammals and are involved in the remodelling of the chromatin of various genes.
- Gamma-activated sequences
-
DNA binding sites specific for signal transducer and activator of transcription 1 (STAT1) homodimers that mediate the response to interferon-γ.
- Interferon-stimulated response elements
-
(ISREs). Common DNA motifs that are bound by interferon-regulatory factors (IRFs). These elements were initially known as IRF enhancers (IRFEs).
Rights and permissions
About this article
Cite this article
Lawrence, T., Natoli, G. Transcriptional regulation of macrophage polarization: enabling diversity with identity. Nat Rev Immunol 11, 750–761 (2011). https://doi.org/10.1038/nri3088
Published:
Issue Date:
DOI: https://doi.org/10.1038/nri3088
This article is cited by
-
MyD88 in myofibroblasts enhances nonalcoholic fatty liver disease-related hepatocarcinogenesis via promoting macrophage M2 polarization
Cell Communication and Signaling (2024)
-
Intestinal epithelial Krüppel-like factor 4 alleviates endotoxemia and atherosclerosis through improving NF-κB/miR-34a-mediated intestinal permeability
Acta Pharmacologica Sinica (2024)
-
Fatty acids metabolism affects the therapeutic effect of anti-PD-1/PD-L1 in tumor immune microenvironment in clear cell renal cell carcinoma
Journal of Translational Medicine (2023)
-
Exploiting E3 ubiquitin ligases to reeducate the tumor microenvironment for cancer therapy
Experimental Hematology & Oncology (2023)
-
Xinyang tablet ameliorates sepsis-induced myocardial dysfunction by regulating Beclin-1 to mediate macrophage autophagy and M2 polarization through LncSICRNT1 targeting E3 ubiquitin ligase TRAF6
Chinese Medicine (2023)