Key Points
-
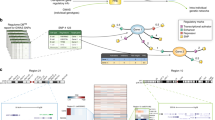
The availability of genetic determinants of multiple sclerosis that have emerged from recent genome-wide association studies creates opportunities to explain for the first time the biological factors that are responsible for the pathophysiology of this disease. These associations implicate genes that fall into two broad categories, immunological genes and neurological genes.
-
Much more work is needed to confirm the disease-associated genetic variants that are responsible for these associations and to attribute this risk to individual loci. In addition, other sorts of genetic variation that have not yet been systematically evaluated in multiple sclerosis, such as rare variants, private mutations and copy number variations, could contribute further to our understanding of heritability.
-
The ultimate proof of causality for a genetic variant will require functional data. This may be partly provided by expression analysis or simple cellular assays but may also require new approaches and animal models that allow exploration of pathway variations using multiple variants or that alter the activity of pathways implicated in disease in humans.
-
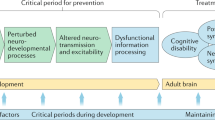
Environmental factors will remain crucial to our understanding of disease risk and pathogenesis. These may be easier to identify if genetics analysis provides clues as to what they may be or if epigenetic modification can be detected that indicates how environmental factors and genes may interact.
-
Other tools, such as experimental medicine in genotyped individuals to detect the biological effects of these polymorphisms as well as modelling and simulation, will also be crucial approaches to dissecting functional roles of these new variants.
Abstract
Susceptibility to multiple sclerosis is jointly determined by genetic and environmental factors, and progress has been made in defining some of these genetic associations, as well as their possible interactions with the environment. However, definitive proof for the involvement of specific genetic determinants in the disease will only come from studies that examine their functional roles in disease pathogenesis. New and combined approaches are needed to analyse the complexity of gene regulation and the functional contribution of each genetic determinant to disease susceptibility or pathophysiology. These studies should proceed in parallel with the use of genetically defined human populations to explore how both genetic and environmental factors affect the function of the pathways in individuals with and without disease, and how these determine the inherited risk of multiple sclerosis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Sospedra, M. & Martin, R. Immunology of multiple sclerosis. Annu. Rev. Immunol. 23, 683–747 (2005).
Lopez-Diego, R. S. & Weiner, H. L. Novel therapeutic strategies for multiple sclerosis — a multifaceted adversary. Nature Rev. Drug Discov. 7, 909–925 (2008).
Correale, J., Fiol, M. & Gilmore, W. The risk of relapses in multiple sclerosis during systemic infections. Neurology 67, 652–659 (2006).
Ascherio, A. & Munger, K. L. Environmental risk factors for multiple sclerosis. Part II: noninfectious factors. Ann. Neurol. 61, 504–513 (2007).
Oksenberg, J. R., Baranzini, S. E., Sawcer, S. & Hauser, S. L. The genetics of multiple sclerosis: SNPs to pathways to pathogenesis. Nature Rev. Genet. 9, 516–526 (2008).
Bertrams, J., Kuwert, E. & Liedtke, U. HL-A antigens and multiple sclerosis. Tissue Antigens 2, 405–408 (1972).
Jersild, C., Svejgaard, A. & Fog, T. HL-A antigens and multiple sclerosis. Lancet 1, 1240–1241 (1972).
Naito, S., Namerow, N., Mickey, M. R. & Terasaki, P. I. Multiple sclerosis: association with HL-A3. Tissue Antigens 2, 1–4 (1972).
Jersild, C. et al. Histocompatibility determinants in multiple sclerosis, with special reference to clinical course. Lancet 2, 1221–1225 (1973).
Lincoln, M. R. et al. A predominant role for the HLA class II region in the association of the MHC region with multiple sclerosis. Nature Genet. 37, 1108–1112 (2005).
Oksenberg, J. R. et al. Mapping multiple sclerosis susceptibility to the HLA-DR locus in African Americans. Am. J. Hum. Genet. 74, 160–167 (2004).
Barcellos, L. F. et al. Heterogeneity at the HLA-DRB1 locus and risk for multiple sclerosis. Hum. Mol. Genet. 15, 2813–2824 (2006).
Dyment, D. A., Ebers, G. C. & Sadovnick, A. D. Genetics of multiple sclerosis. Lancet Neurol. 3, 104–110 (2004).
Fogdell-Hahn, A., Ligers, A., Gronning, M., Hillert, J. & Olerup, O. Multiple sclerosis: a modifying influence of HLA class I genes in an HLA class II associated autoimmune disease. Tissue Antigens 55, 140–148 (2000).
Harbo, H. F. et al. Genes in the HLA class I region may contribute to the HLA class II-associated genetic susceptibility to multiple sclerosis. Tissue Antigens 63, 237–247 (2004).
Brynedal, B. et al. HLA-A confers an HLA-DRB1 independent influence on the risk of multiple sclerosis. PLoS ONE 2, e664 (2007).
Yeo, T. W. et al. A second major histocompatibility complex susceptibility locus for multiple sclerosis. Ann. Neurol. 61, 228–236 (2007).
Hafler, D. A. et al. Risk alleles for multiple sclerosis identified by a genome-wide study. N. Engl. J. Med. 357, 851–862 (2007). This study shows that multiple sclerosis is associated with several genetic variants in or around immunologically relevant genes. The MHC region confers the largest risk, whereas the contributions from other risk genes, such as IL7R and IL2R , are small by comparison.
Lundmark, F. et al. Variation in interleukin 7 receptor α chain (IL7R) influences risk of multiple sclerosis. Nature Genet. 39, 1108–1113 (2007).
Ban, M. et al. Replication analysis identifies TYK2 as a multiple sclerosis susceptibility factor. Eur. J. Hum. Genet. 18 Mar 2009 (doi:10.1038/ejhg.2009.41).
Reich, D. et al. A whole-genome admixture scan finds a candidate locus for multiple sclerosis susceptibility. Nature Genet. 37, 1113–1118 (2005).
Hafler, J. P. et al. CD226 Gly307Ser association with multiple autoimmune diseases. Genes Immun. 10, 305–310 (2009).
Bernardinelli, L. et al. Association between the ACCN1 gene and multiple sclerosis in central east Sardinia. PLoS ONE 2, e480 (2007).
Aulchenko, Y. S. et al. Genetic variation in the KIF1B locus influences susceptibility to multiple sclerosis. Nature Genet. 40, 1402–1403 (2008).
Wemmie, J. A., Price, M. P. & Welsh, M. J. Acid-sensing ion channels: advances, questions and therapeutic opportunities. Trends Neurosci. 29, 578–586 (2006).
Boldogh, I. R. & Pon, L. A. Mitochondria on the move. Trends Cell Biol. 17, 502–510 (2007).
Nangaku, M. et al. KIF1B, a novel microtubule plus end-directed monomeric motor protein for transport of mitochondria. Cell 79, 1209–1220 (1994).
Donnelly, P. Progress and challenges in genome-wide association studies in humans. Nature 456, 728–731 (2008).
Vavouri, T., McEwen, G. K., Woolfe, A., Gilks, W. R. & Elgar, G. Defining a genomic radius for long-range enhancer action: duplicated conserved non-coding elements hold the key. Trends Genet. 22, 5–10 (2006).
Feuk, L., Carson, A. R. & Scherer, S. W. Structural variation in the human genome. Nature Rev. Genet. 7, 85–97 (2006).
Nejentsev, S., Walker, N., Riches, D., Egholm, M. & Todd, J. A. Rare variants of IFIH1, a gene implicated in antiviral responses, protect against type 1 diabetes. Science 5 Mar 2009 (doi:10.1126/science.1167728). This paper shows that re-sequencing studies can pinpoint disease-causing genes in genomic regions initially identified by genome-wide association studies.
Madsen, L. S. et al. A humanized model for multiple sclerosis using HLA-DR2 and a human T-cell receptor. Nature Genet. 23, 343–347 (1999).
Molberg, O. et al. Tissue transglutaminase selectively modifies gliadin peptides that are recognized by gut-derived T cells in celiac disease. Nature Med. 4, 713–717 (1998).
Cookson, W., Liang, L., Abecasis, G., Moffatt, M. & Lathrop, M. Mapping complex disease traits with global gene expression. Nature Rev. Genet. 10, 184–194 (2009).
Chen, Y. et al. Variations in DNA elucidate molecular networks that cause disease. Nature 452, 429–435 (2008).
Emilsson, V. et al. Genetics of gene expression and its effect on disease. Nature 452, 423–428 (2008). This study assesses the relationship between DNA sequence variants and gene expression. For a recent review on this subject see reference 34.
Goring, H. H. et al. Discovery of expression QTLs using large-scale transcriptional profiling in human lymphocytes. Nature Genet. 39, 1208–1216 (2007).
Moffatt, M. F. et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature 448, 470–473 (2007).
Maier, L. M. et al. Soluble IL-2RA levels in multiple sclerosis subjects and the effect of soluble IL-2RA on immune responses. J. Immunol. 182, 1541–1547 (2009).
Maier, L. M. et al. IL2RA genetic heterogeneity in multiple sclerosis and type 1 diabetes susceptibility and soluble interleukin-2 receptor production. PLoS Genet. 5, e1000322 (2009).
De Jager, P. L. et al. The role of the CD58 locus in multiple sclerosis. Proc. Natl Acad. Sci. USA 106, 5264–5269 (2009).
Gregory, S. G. et al. Interleukin 7 receptor α chain (IL7R) shows allelic and functional association with multiple sclerosis. Nature Genet. 39, 1083–1091 (2007).
Nunnari, G. et al. Exogenous IL-7 induces Fas-mediated human neuronal apoptosis: potential effects during human immunodeficiency virus type 1 infection. J. Neurovirol. 11, 319–328 (2005).
Park, I. H. et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature 451, 141–146 (2008).
Takahashi, K. et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872 (2007).
Yu, J. et al. Induced pluripotent stem cell lines derived from human somatic cells. Science 318, 1917–1920 (2007).
Aasen, T. et al. Efficient and rapid generation of induced pluripotent stem cells from human keratinocytes. Nature Biotechnol. 26, 1276–1284 (2008).
Loh, Y. H. et al. Generation of induced pluripotent stem cells from human blood. Blood 18 Mar 2009 (doi:10.1182/blood-2009-02-204800).
Kaji, K. et al. Virus-free induction of pluripotency and subsequent excision of reprogramming factors. Nature 58, 771–775 (2009).
Yamanaka, S. A fresh look at iPS cells. Cell 137, 13–17 (2009). This is mandatory reading for those wishing to catch up with the iPS cell field.
Dimos, J. T. et al. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science 321, 1218–1221 (2008).
Ebert, A. D. et al. Induced pluripotent stem cells from a spinal muscular atrophy patient. Nature 457, 277–280 (2009). This is the first study to show that human iPS cells can be used to model the specific pathology seen in a genetically inherited disease.
Soldner, F. et al. Parkinson's disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell 136, 964–977 (2009).
Borrelli, E., Nestler, E. J., Allis, C. D. & Sassone-Corsi, P. Decoding the epigenetic language of neuronal plasticity. Neuron 60, 961–974 (2008).
Chao, M. J. et al. Epigenetics in multiple sclerosis susceptibility: difference in transgenerational risk localizes to the major histocompatibility complex. Hum. Mol. Genet. 18, 261–266 (2009).
Zhang, X. et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in arabidopsis. Cell 126, 1189–1201 (2006).
Mockler, T. C. et al. Applications of DNA tiling arrays for whole-genome analysis. Genomics 85, 1–15 (2005).
Shen, S. et al. Age-dependent epigenetic control of differentiation inhibitors is critical for remyelination efficiency. Nature Neurosci. 11, 1024–1034 (2008). References 54 and 58 suggest that investigating dysregulated posttranslational modifications in multiple sclerosis may contribute to our understanding of its pathogenesis.
Friese, M. A. et al. The value of animal models for drug development in multiple sclerosis. Brain 129, 1940–1952 (2006).
Steinman, L. Blocking adhesion molecules as therapy for multiple sclerosis: natalizumab. Nature Rev. Drug Discov. 4, 510–518 (2005).
Gregersen, J. W. et al. Functional epistasis on a common MHC haplotype associated with multiple sclerosis. Nature 443, 574–577 (2006).
Caillier, S. J. et al. Uncoupling the roles of HLA-DRB1 and HLA-DRB5 genes in multiple sclerosis. J. Immunol. 181, 5473–5480 (2008).
Polman, C. H. et al. A randomized, placebo-controlled trial of natalizumab for relapsing multiple sclerosis. N. Engl. J. Med. 354, 899–910 (2006).
Coles, A. J. et al. Alemtuzumab vs. interferon β-1a in early multiple sclerosis. N. Engl. J. Med. 359, 1786–1801 (2008).
Friese, M. A. et al. Opposing effects of HLA class I molecules in tuning autoreactive CD8+ T cells in multiple sclerosis. Nature Med. 14, 1227–1235 (2008). This study exemplifies how functional genetics can be used to understand disease-association studies that are directly relevant to multiple sclerosis.
Bell, G. I., Horita, S. & Karam, J. H. A polymorphic locus near the human insulin gene is associated with insulin-dependent diabetes mellitus. Diabetes 33, 176–183 (1984).
Vafiadis, P. et al. Insulin expression in human thymus is modulated by INS VNTR alleles at the IDDM2 locus. Nature Genet. 15, 289–292 (1997).
Pugliese, A. et al. The insulin gene is transcribed in the human thymus and transcription levels correlated with allelic variation at the INS VNTR-IDDM2 susceptibility locus for type 1 diabetes. Nature Genet. 15, 293–297 (1997).
Chentoufi, A. A. & Polychronakos, C. Insulin expression levels in the thymus modulate insulin-specific autoreactive T-cell tolerance: the mechanism by which the IDDM2 locus may predispose to diabetes. Diabetes 51, 1383–1390 (2002).
Lunemann, J. D. et al. EBNA1-specific T cells from patients with multiple sclerosis cross react with myelin antigens and co-produce IFN-γ and IL-2. J. Exp. Med. 205, 1763–1773 (2008).
Harkiolaki, M. et al. T cell-mediated autoimmune disease due to low-affinity crossreactivity to common microbial peptides. Immunity 30, 348–357 (2009).
Oldstone, M. B. Molecular mimicry and autoimmune disease. Cell 50, 819–820 (1987).
Quintana, F. J. et al. Control of Treg and TH17 cell differentiation by the aryl hydrocarbon receptor. Nature 453, 65–71 (2008).
Veldhoen, M. et al. The aryl hydrocarbon receptor links TH17-cell-mediated autoimmunity to environmental toxins. Nature 453, 106–109 (2008). References 73 and 74 show the role of the environmental toxin dioxin in affecting T Reg and T H 17 cell activation and provide a potentially useful clue about the possible role of environmental triggers in disease initiation.
Bettelli, E., Korn, T., Oukka, M. & Kuchroo, V. K. Induction and effector functions of T H17 cells. Nature 453, 1051–1057 (2008).
Tzartos, J. S. et al. Interleukin-17 production in central nervous system-infiltrating T cells and glial cells is associated with active disease in multiple sclerosis. Am. J. Pathol. 172, 146–155 (2008).
Ramagopalan, S. V. et al. Expression of the multiple sclerosis-associated MHC class II allele HLA-DRB1*1501 is regulated by vitamin D. PLoS Genet. 5, e1000369 (2009).
Friese, M. A. et al. Acid-sensing ion channel-1 contributes to axonal degeneration in autoimmune inflammation of the central nervous system. Nature Med. 13, 1483–1489 (2007).
Gimeno, R. et al. Monitoring the effect of gene silencing by RNA interference in human CD34+ cells injected into newborn RAG2−/− γc−/− mice: functional inactivation of p53 in developing T cells. Blood 104, 3886–3893 (2004).
Traggiai, E. et al. Development of a human adaptive immune system in cord blood cell-transplanted mice. Science 304, 104–107 (2004).
Shultz, L. D., Ishikawa, F. & Greiner, D. L. Humanized mice in translational biomedical research. Nature Rev. Immunol. 7, 118–130 (2007).
Baranzini, S. E. et al. Pathway and network-based analysis of genome-wide association studies in multiple sclerosis. Hum. Mol. Genet. 13 Mar 2009 (doi:10.1093/hmg/ddp120).
Baranzini, S. E. et al. Genome-wide association analysis of susceptibility and clinical phenotype in multiple sclerosis. Hum. Mol. Genet. 18, 767–778 (2009).
McFarland, H. F. & Martin, R. Multiple sclerosis: a complicated picture of autoimmunity. Nature Immunol. 8, 913–919 (2007).
Frischer, J. M. et al. The relation between inflammation and neurodegeneration in multiple sclerosis brains. Brain 31 Mar 2009 (doi:10.1093/brain/awp070).
Frohman, E. M., Racke, M. K. & Raine, C. S. Multiple sclerosis — the plaque and its pathogenesis. N. Engl. J. Med. 354, 942–955 (2006).
Goodnow, C. C. Multistep pathogenesis of autoimmune disease. Cell 130, 25–35 (2007).
Acknowledgements
We thank N. Willcox and A. Vincent for critical reading of the manuscript. Work in the authors' laboratories is supported by the Danish and UK Medical Research Councils, the Karen Elise Jensen Foundation, the Lundbeck Foundation, the Danish Multiple Sclerosis Society, the European Union (European Commission Descartes Prize, FP6 (Neuropromise, Mugen and ARDIS) and FP7 (SYBILLA)). M.A.F. is supported by the DFG Emmy Noether Programme (grant number FR1720/3-1).
Author information
Authors and Affiliations
Corresponding authors
Related links
Related links
OMIM
FURTHER INFORMATION
Glossary
- Demyelination
-
Damage to the myelin sheath surrounding nerves in the brain and spinal cord, which affects the function of the nerves involved.
- Axonal degeneration
-
Loss of nerve fibres in response to local damage.
- Genome-wide association study
-
A study designed to look for association between disease and a dense set of markers covering the entire genome.
- Linkage disequilibrium
-
A situation in which alleles in a chromosomal region occur together more often than can be accounted for by chance, indicating that the alleles are in close proximity on the DNA strand and are most likely to be passed on together within a population.
- Candidate gene association study
-
A study that compares the allele frequency of a gene for which there is evidence, usually functional, for a possible role in a disease or trait of interest in cases and controls to assess the contribution of genetic variants to phenotypes in specific populations.
- HLA-DR2 haplotype
-
A combination of alleles at many linked loci that are inherited together; in this case the MHC class II alleles HLA-DRB1*1501 and HLA-DRB5*0101.
- Single nucleotide polymorphism
-
(SNP). Genomic variant in which a single base in the DNA differs from the usual base at that position. SNPs are the most common type of variation in the human genome.
- Private variant
-
The specific genetic variant that only occurs in one individual or family that functionally gives rise to an increased risk conferred by the causal gene or genomic region.
- Next generation sequencing technology
-
Technology that allows for parallel sequencing of massive amounts of DNA. This technology can be used for deep sequencing to sequence whole genomes, transcriptome analysis and for the identification of rare mutants.
- eQTL mapping
-
Combination of quantitative trait loci (regions of DNA that are closely linked to a phenotypic outcome) mapping and gene-expression analysis to study the genetic basis of gene expression and, by extension, biological regulation.
- Regulatory T cell
-
(TReg cell). A type of CD4+ T cell that is characterized by its expression of forkhead box P3 and high levels of CD25. TReg cells can downmodulate many types of immune response.
- Induced pluripotent stem cell
-
A type of pluripotent stem cell that is artificially derived from a non-pluripotent cell, typically an adult somatic cell, by retroviral transfer of a panel of developmentally regulated genes. They can differentiate into multiple cell lineages.
- Epigenome
-
The chromatin states that are found along the whole genome, defined for a given time point and cell type.
- Epistatic interaction
-
Any non-additive interaction between two or more variants at different loci, such that their combined effect on a phenotype differs from the one that would be produced if the two genes were acting independently.
- T helper 17
-
A subset of CD4+ T helper cells that produce interleukin-17 (IL-17) and that are thought to be important in inflammatory and autoimmune diseases. Their generation involves transforming growth factor-β, IL-6, IL-23 or IL-21, IL-1 and the transcription factors RORγt and STAT3.
- Tissue acidosis
-
Lowered pH (acidosis) that is caused by increased glycolysis, production of lactic acid and decreased extracellular and intracellular pH. Tissue acidosis is associated with imbalance between energy supply and demand.
Rights and permissions
About this article
Cite this article
Fugger, L., Friese, M. & Bell, J. From genes to function: the next challenge to understanding multiple sclerosis. Nat Rev Immunol 9, 408–417 (2009). https://doi.org/10.1038/nri2554
Issue Date:
DOI: https://doi.org/10.1038/nri2554
This article is cited by
-
Antigen recognition detains CD8+ T cells at the blood-brain barrier and contributes to its breakdown
Nature Communications (2023)
-
Magnetically guided virus stamping for the targeted infection of single cells or groups of cells
Nature Protocols (2019)
-
Nucleolar TRF2 attenuated nucleolus stress-induced HCC cell-cycle arrest by altering rRNA synthesis
Cell Death & Disease (2018)
-
Neural Stem Cell-Based Regenerative Approaches for the Treatment of Multiple Sclerosis
Molecular Neurobiology (2018)
-
Optimized nanoparticle-mediated delivery of CRISPR-Cas9 system for B cell intervention
Nano Research (2018)