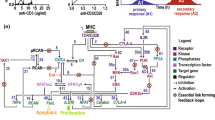
Key Points
-
Recent experimental evidence resolves two important gaps in our understanding of the molecular mechanisms by which antigen receptors signal to activate nuclear factor-κB (NF-κB). This new information includes: first, how protein kinase C (PKC) isoforms directly activate the CARMA1 (caspase-recruitment domain (CARD)–membrane-associated guanylate kinase (MAGUK) protein 1) signalosome; and second, how specific signalling molecules that also promote NF-κB activation downstream of innate immune receptors participate in this pathway following the recruitment of MALT1 (mucosa-associated-lymphoid-tissue lymphoma-translocation gene 1).
-
Antigen-receptor stimulation initiates a proximal tyrosine-phosphorylation cascade that promotes activation of phospholipase C-γ isoforms, leading to the production of second messengers that are essential for PKC activation. Activated PKCβ (in B cells) and PKCθ (in T cells) subsequently phosphorylate key serine residues in the PKC-regulated domain (PRD) of CARMA1.
-
In unstimulated cells, CARMA1 assumes a closed conformation through intramolecular interactions. PKC-mediated phosphorylation of the PRD of CARMA1 triggers a conformational change that exposes downstream CARMA1 protein-binding domains required for signalosome assembly, including: the CARD required for B-cell lymphoma 10 (BCL-10) binding, the coiled-coil (CC) domain for CARMA1 oligomerization, and the GUK domain, which is predicted to mediate higher-order multimerization of oligomerized CARMA1 complexes.
-
Activated CARMA1 initiates an oligomerization cascade of BCL-10 and MALT1, which is followed by the recruitment of trimerized TRAF2 (tumour-necrosis factor receptor (TNFR)-associated factor 2) or TRAF6. On the basis of studies of innate immune-receptor signalling, TRAF2 or TRAF6 oligomerization stimulates TRAF-mediated K63-linked polyubiquitylation of target proteins. These K63-linked polyubiquitylated targets then function as scaffolds that recruit ubiquitin-binding proteins associated with TAK1 (transforming-growth-factor-β-activated kinase 1) and IKK (inhibitor of nuclear factor-κB (IκB) kinase) complexes into the CARMA1 signalosome.
-
The specific and non-redundant role for CARMA1 in antigen-receptor-mediated activation of NF-κB makes this protein an appealing target for pharmacological regulation in a range of specific immune disorders, as well in human B-cell lymphomas characterized by dysregulated NF-κB activation.
Abstract
The recognition of antigen by B- or T-cell receptors initiates an intracellular signalling cascade that results in the nuclear translocation and activation of the transcription factor nuclear factor-κB (NF-κB). NF-κB is an important regulator of lymphocyte development and function, and its dysregulation is associated with many immune disorders. Defining the mechanisms that transmit signals from the antigen receptor to NF-κB is therefore an important goal for immunologists. In this Review, we merge information gleaned from research of the innate immune system with what we know about antigen-receptor signals in the adaptive immune system, to propose a cohesive model of how antigen receptors activate NF-κB.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Silverman, N. & Maniatis, T. NF-κB signaling pathways in mammalian and insect innate immunity. Genes Dev. 15, 2321–2342 (2001).
Flajnik, M. F. & Du Pasquier, L. Evolution of innate and adaptive immunity: can we draw a line? Trends Immunol. 25, 640–644 (2004).
Litman, G. W., Cannon, J. P. & Dishaw, L. J. Reconstructing immune phylogeny: new perspectives. Nature Rev. Immunol. 5, 866–879 (2005).
Magor, B. G. & Magor, K. E. Evolution of effectors and receptors of innate immunity. Dev. Comp. Immunol. 25, 651–682 (2001).
Sommer, K. et al. Phosphorylation of the CARMA1 linker controls NF-κB activation. Immunity 23, 561–574 (2005).
Matsumoto, R. et al. Phosphorylation of CARMA1 plays a critical role in T-cell receptor-mediated NF-κB activation. Immunity 23, 575–585 (2005). References 5 and 6 establish that CARMA1 is the direct downstream target of PKC, and that PKC phosphorylation triggers the activation of CARMA1.
Shinohara, H. et al. PKCβ regulates BCR-mediated IKK activation by facilitating the interaction between TAK1 and CARMA1. J. Exp. Med. 202, 1423–1431 (2005). Reference 7, together with references 10–12, verifies a role for TAK1 in BCR and TCR signalling using a genetically altered B-cell line (reference 7), or mouse models with conditional genetic deletions of Tak1 in B cells (reference 10) or T cells (references 11 and 12).
Zhou, H. et al. Bcl10 activates the NF-κB pathway through ubiquitination of NEMO. Nature 427, 167–171 (2004).
Sun, L., Deng, L., Ea, C. K., Xia, Z. P. & Chen, Z. J. The TRAF6 ubiquitin ligase and TAK1 kinase mediate IKK activation by BCL10 and MALT1 in T lymphocytes. Mol. Cell 14, 289–301 (2004). References 8 and 9 describe the original findings that activating ubiquitylation events transmit antigen-receptor signals downstream of MALT1.
Sato, S. et al. Essential function for the kinase TAK1 in innate and adaptive immune responses. Nature Immunol. 6, 1087–1095 (2005).
Wan, Y. Y., Chi, H., Xie, M., Schneider, M. D. & Flavell, R. A. The kinase TAK1 integrates antigen and cytokine receptor signaling for T–cell development, survival and function. Nature Immunol. 7, 851–858 (2006).
Liu, H. H., Xie, M., Schneider, M. D. & Chen, Z. J. Essential role of TAK1 in thymocyte development and activation. Proc. Natl Acad. Sci. USA 103, 11677–11682 (2006).
Siebenlist, U., Brown, K. & Claudio, E. Control of lymphocyte development by nuclear factor-κB. Nature Rev. Immunol. 5, 435–445 (2005).
Dal Porto, J. M. et al. B-cell antigen receptor signaling 101. Mol. Immunol. 41, 599–613 (2004).
Kurosaki, T. et al. Regulation of the phospholipase C-γ2 pathway in B cells. Immunol. Rev. 176, 19–29 (2000).
van Leeuwen, J. E. & Samelson, L. E. T-cell antigen-receptor signal transduction. Curr. Opin. Immunol. 11, 242–248 (1999).
Weil, R. & Israel, A. T-cell-receptor- and B-cell-receptor-mediated activation of NF-κB in lymphocytes. Curr. Opin. Immunol. 16, 374–381 (2004).
Sun, Z. et al. PKC-θ is required for TCR-induced NF-κB activation in mature but not immature T lymphocytes. Nature 404, 402–407 (2000).
Su, T. T. et al. PKC-β controls IκB kinase lipid raft recruitment and activation in response to BCR signaling. Nature Immunol. 3, 780–786 (2002).References 18 and 19 show that PKCθ (in T cells) and PKCβ (in B cells) have crucial and non-redundant roles in the antigen-receptor signalling pathways that activate NF-κB.
Thome, M. CARMA1, BCL-10 and MALT1 in lymphocyte development and activation. Nature Rev. Immunol. 4, 348–359 (2004).
Hara, H. et al. The MAGUK family protein CARD11 is essential for lymphocyte activation. Immunity 18, 763–775 (2003).
Newton, K. & Dixit, V. M. Mice lacking the CARD of CARMA1 exhibit defective B-lymphocyte development and impaired proliferation of their B and T lymphocytes. Curr. Biol. 13, 1247–1251 (2003).
Egawa, T. et al. Requirement for CARMA1 in antigen receptor-induced NF-κB activation and lymphocyte proliferation. Curr. Biol. 13, 1252–1258 (2003).
Jun, J. E. et al. Identifying the MAGUK protein Carma-1 as a central regulator of humoral immune responses and atopy by genome-wide mouse mutagenesis. Immunity 18, 751–762 (2003). References 21–24 describe the consequences of CARMA1 deficiency in primary lymphocytes using mouse models, including both genetic deletion and mutations that cause a loss of CARMA1 function.
Wang, D. et al. A requirement for CARMA1 in TCR-induced NF-κB activation. Nature Immunol. 3, 830–835 (2002).
Gaide, O. et al. CARMA1 is a critical lipid raft-associated regulator of TCR-induced NF-κB activation. Nature Immunol. 3, 836–843 (2002).
Pomerantz, J. L., Denny, E. M. & Baltimore, D. CARD11 mediates factor-specific activation of NF-κB by the T-cell receptor complex. EMBO J. 21, 5184–5194 (2002).
Gaide, O. et al. Carma1, a CARD-containing binding partner of Bcl10, induces Bcl10 phosphorylation and NF-κB activation. FEBS Lett. 496, 121–127 (2001).
Bertin, J. et al. CARD11 and CARD14 are novel caspase recruitment domain (CARD)/membrane-associated guanylate kinase (MAGUK) family members that interact with BCL10 and activate NF-κB. J. Biol. Chem. 276, 11877–11882 (2001).
McAllister-Lucas, L. M. et al. Bimp1, a MAGUK family member linking protein kinase C activation to Bcl10-mediated NF-κB induction. J. Biol. Chem. 276, 30589–30597 (2001).
McGee, A. W. et al. Structure of the SH3-guanylate kinase module from PSD-95 suggests a mechanism for regulated assembly of MAGUK scaffolding proteins. Mol. Cell 8, 1291–1301 (2001).
Zhang, Q. et al. Inactivating mutations and overexpression of BCL10, a caspase recruitment domain-containing gene, in MALT lymphoma with t(1;14)(p22;q32). Nature Genet. 22, 63–68 (1999).
Willis, T. G. et al. Bcl10 is involved in t(1;14)(p22;q32) of MALT B-cell lymphoma and mutated in multiple tumor types. Cell 96, 35–45 (1999).
Ruefli-Brasse, A. A., French, D. M. & Dixit, V. M. Regulation of NF-κB-dependent lymphocyte activation and development by paracaspase. Science 302, 1581–1584 (2003). References 34 and 48 highlight the phenotypic abnormalities of lymphocytes from mice with genetic deletion of the Malt1 locus.
Deng, L. et al. Activation of the IκB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell 103, 351–361 (2000). This paper describes seminal experiments establishing K63-linked polyubiquitylation as a key mechanism for regulating proteins in the signalling pathway of NF-κB activation downstream of TRAF6.
Ruland, J. et al. Bcl10 is a positive regulator of antigen receptor-induced activation of NF-κB and neural tube closure. Cell 104, 33–42 (2001).
Xue, L. et al. Defective development and function of Bcl10-deficient follicular, marginal zone and B1 B cells. Nature Immunol. 4, 857–865 (2003). References 36 and 37 identify the essential role of BCL-10 in antigen-receptor signalling in primary lymphocytes, using knockout mouse models.
Koseki, T. et al. CIPER, a novel NF-κB-activating protein containing a caspase recruitment domain with homology to herpesvirus-2 protein E10. J. Biol. Chem. 274, 9955–9961 (1999).
Srinivasula, S. M. et al. CLAP, a novel caspase recruitment domain-containing protein in the tumor necrosis factor receptor pathway, regulates NF-κB activation and apoptosis. J. Biol. Chem. 274, 17946–17954 (1999).
Hara, H. et al. The molecular adapter Carma1 controls entry of IκB kinase into the central immune synapse. J. Exp. Med. 200, 1167–1177 (2004).
Wang, D. et al. CD3/CD28 costimulation-induced NF-κB activation is mediated by recruitment of protein kinase C-θ, Bcl10 and IκB kinase-β to the immunological synapse through CARMA1. Mol. Cell. Biol. 24, 164–171 (2004).
Guiet, C. & Vito, P. Caspase recruitment domain (CARD)-dependent cytoplasmic filaments mediate Bcl10-induced NF-κB activation. J. Cell Biol. 148, 1131–1140 (2000).
Ruefli-Brasse, A. A., Lee, W. P., Hurst, S. & Dixit, V. M. Rip2 participates in Bcl10 signaling and T-cell receptor-mediated NF-κB activation. J. Biol. Chem. 279, 1570–1574 (2004).
Lucas, P. C. et al. Bcl10 and MALT1, independent targets of chromosomal translocation in malt lymphoma, cooperate in a novel NF-κB signaling pathway. J. Biol. Chem. 276, 19012–19019 (2001).
Dierlamm, J. et al. The apoptosis inhibitor gene API2 and a novel 18q gene, MLT, are recurrently rearranged in the t(11;18)(q21;q21) associated with mucosa-associated lymphoid tissue lymphomas. Blood 93, 3601–3609 (1999).
Morgan, J. A. et al. Breakpoints of the t(11;18)(q21;q21) in mucosa-associated lymphoid tissue (MALT) lymphoma lie within or near the previously undescribed gene MALT1 in chromosome 18. Cancer Res. 59, 6205–6213 (1999).
Uren, A. G. et al. Identification of paracaspases and metacaspases: two ancient families of caspase-like proteins, one of which plays a key role in MALT lymphoma. Mol. Cell 6, 961–967 (2000).
Ruland, J., Duncan, G. S., Wakeham, A. & Mak, T. W. Differential requirement for Malt1 in T- and B-cell antigen receptor signaling. Immunity 19, 749–758 (2003).
Wegener, E. et al. Essential role for IκB kinase-β in remodeling Carma1–Bcl10–Malt1 complexes upon T-cell activation. Mol. Cell 23, 13–23 (2006).
Park, Y. C., Burkitt, V., Villa, A. R., Tong, L. & Wu, H. Structural basis for self-association and receptor recognition of human TRAF2. Nature 398, 533–538 (1999).
Ye, H. & Wu, H. Thermodynamic characterization of the interaction between TRAF2 and tumor necrosis factor receptor peptides by isothermal titration calorimetry. Proc. Natl Acad. Sci. USA 97, 8961–8966 (2000). References 50 and 51 together provide strong structural evidence for TRAF trimerization, and provide a model of TRAF recruitment to upstream receptors on the basis of thermodynamic data.
Baud, V. et al. Signaling by proinflammatory cytokines: oligomerization of TRAF2 and TRAF6 is sufficient for JNK and IKK activation and target gene induction via an amino-terminal effector domain. Genes Dev. 13, 1297–1308 (1999).
Chen, Z. J., Parent, L. & Maniatis, T. Site-specific phosphorylation of IκBα by a novel ubiquitination-dependent protein kinase activity. Cell 84, 853–862 (1996).
Stancovski, I. & Baltimore, D. NF-κB activation: the IκB kinase revealed? Cell 91, 299–302 (1997).
Maniatis, T. Catalysis by a multiprotein IκB kinase complex. Science 278, 818–819 (1997).
Hofmann, R. M. & Pickart, C. M. Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell 96, 645–653 (1999).
Schnell, J. D. & Hicke, L. Non-traditional functions of ubiquitin and ubiquitin-binding proteins. J. Biol. Chem. 278, 35857–35860 (2003).
Yamaguchi, K. et al. Identification of a member of the MAPKKK family as a potential mediator of TGFβ signal transduction. Science 270, 2008–2011 (1995).
Ninomiya-Tsuji, J. et al. The kinase TAK1 can activate the NIK–IκB as well as the MAP kinase cascade in the IL-1 signalling pathway. Nature 398, 252–256 (1999).
Irie, T., Muta, T. & Takeshige, K. TAK1 mediates an activation signal from Toll-like receptor(s) to nuclear factor-κB in lipopolysaccharide-stimulated macrophages. FEBS Lett. 467, 160–164 (2000).
Lee, J., Mira-Arbibe, L. & Ulevitch, R. J. TAK1 regulates multiple protein kinase cascades activated by bacterial lipopolysaccharide. J. Leukocyte Biol. 68, 909–915 (2000).
Wang, C. et al. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature 412, 346–351 (2001). Using elegant biochemical techniques, the authors identify TAK1 as the key IKK kinase downstream of TRAF6, and show that TAK1 phosphorylates and activates MKK6 also.
Shim, J. H. et al. TAK1, but not TAB1 or TAB2, plays an essential role in multiple signaling pathways in vivo. Genes Dev. 19, 2668–2681 (2005).
Takaesu, G. et al. TAB2, a novel adaptor protein, mediates activation of TAK1 MAPKKK by linking TAK1 to TRAF6 in the IL-1 signal transduction pathway. Mol. Cell 5, 649–658 (2000).
Jiang, Z., Ninomiya-Tsuji, J., Qian, Y., Matsumoto, K. & Li, X. Interleukin-1 (IL-1) receptor-associated kinase-dependent IL-1-induced signaling complexes phosphorylate TAK1 and TAB2 at the plasma membrane and activate TAK1 in the cytosol. Mol. Cell Biol. 22, 7158–7167 (2002).
Cheung, P. C., Nebreda, A. R. & Cohen, P. TAB3, a new binding partner of the protein kinase TAK1. Biochem. J. 378, 27–34 (2004).
Shibuya, H. et al. TAB1: an activator of the TAK1 MAPKKK in TGFβ signal transduction. Science 272, 1179–1182 (1996).
Sakurai, H., Miyoshi, H., Toriumi, W. & Sugita, T. Functional interactions of transforming growth factor-β-activated kinase 1 with IκB kinases to stimulate NF-κB activation. J. Biol. Chem. 274, 10641–10648 (1999).
Kishimoto, K., Matsumoto, K. & Ninomiya-Tsuji, J. TAK1 mitogen-activated protein kinase kinase kinase is activated by autophosphorylation within its activation loop. J. Biol. Chem. 275, 7359–7364 (2000).
Singhirunnusorn, P., Suzuki, S., Kawasaki, N., Saiki, I. & Sakurai, H. Critical roles of threonine 187 phosphorylation in cellular stress-induced rapid and transient activation of transforming growth factor-β-activated kinase 1 (TAK1) in a signaling complex containing TAK1-binding protein TAB1 and TAB2. J. Biol. Chem. 280, 7359–7368 (2005).
Ishitani, T. et al. Role of the TAB2-related protein TAB3 in IL-1 and TNF signaling. EMBO J. 22, 6277–6288 (2003).
Takaesu, G. et al. TAK1 is critical for IκB kinase-mediated activation of the NF-κB pathway. J. Mol. Biol. 326, 105–115 (2003).
Kanayama, A. et al. TAB2 and TAB3 activate the NF-κB pathway through binding to polyubiquitin chains. Mol. Cell 15, 535–548 (2004). In this paper, the importance of TAB2 and TAB3 in the function of TAK1 complexes is established. This is also the first description of a K63-ubiquitin-binding domain used by molecules activating the NF-κB pathway, which indicates that K63-linked polyubiquitin chains regulate this pathway by modulating protein interactions.
Meylan, E. & Tschopp, J. The RIP kinases: crucial integrators of cellular stress. Trends Biochem. Sci. 30, 151–159 (2005).
Yoneda, T. et al. Regulatory mechanisms of TRAF2-mediated signal transduction by Bcl10, a MALT lymphoma-associated protein. J. Biol. Chem. 275, 11114–11120 (2000).
Hosokawa, Y., Suzuki, H., Suzuki, Y., Takahashi, R. & Seto, M. Antiapoptotic function of apoptosis inhibitor 2–MALT1 fusion protein involved in t(11;18)(q21;q21) mucosa-associated lymphoid tissue lymphoma. Cancer Res. 64, 3452–3457 (2004).
Agou, F. et al. The trimerization domain of NEMO is composed of the interacting C-terminal CC2 and LZ coiled-coil subdomains. J. Biol. Chem. 279, 27861–27869 (2004). In this paper, evidence for a novel model of IKKγ structural folding and trimerization is described.
Zandi, E., Chen, Y. & Karin, M. Direct phosphorylation of IκB by IKKα and IKKβ: discrimination between free and NF-κB-bound substrate. Science 281, 1360–1363 (1998).
Yamaoka, S. et al. Complementation cloning of NEMO, a component of the IκB kinase complex essential for NF-κB activation. Cell 93, 1231–1240 (1998).
Rothwarf, D. M., Zandi, E., Natoli, G. & Karin, M. IKKγ is an essential regulatory subunit of the IκB kinase complex. Nature 395, 297–300 (1998).
Stilo, R. et al. Physical and functional interaction of CARMA1 and CARMA3 with Iκkinase-γ–NFκB essential modulator. J. Biol. Chem. 279, 34323–34331 (2004).
Ea, C. K., Deng, L., Xia, Z. P., Pineda, G. & Chen, Z. J. Activation of IKK by TNFα requires site-specific ubiquitination of RIP1 and polyubiquitin binding by NEMO. Mol. Cell 22, 245–257 (2006).
Wu, C. J., Conze, D. B., Li, T., Srinivasula, S. M. & Ashwell, J. D. NEMO is a sensor of Lys63-linked polyubiquitination and functions in NF-κB activation. Nature Cell Biol. 8, 398–406 (2006). References 82 and 83 each describe the requirement of IKKγ binding to K63-linked polyubiquitin chains of activated RIP1 for NF-κB activation downstream of TNF-receptor signals.
Tang, E. D., Wang, C. Y., Xiong, Y. & Guan, K. L. A role for NF-κB essential modifier/IκB kinase-γ (NEMO/IKKγ) ubiquitination in the activation of the IκB kinase complex by tumor necrosis factor-α. J. Biol. Chem. 278, 37297–37305 (2003).
Scharschmidt, E., Wegener, E., Heissmeyer, V., Rao, A. & Krappmann, D. Degradation of Bcl10 induced by T-cell activation negatively regulates NF-κB signaling. Mol. Cell Biol. 24, 3860–3873 (2004).
Hu, S. et al. cIAP2 is a ubiquitin protein ligase for BCL10 and is dysregulated in mucosa-associated lymphoid tissue lymphomas. J. Clin. Invest. 116, 174–181 (2006).
Perry, W. L. et al. The itchy locus encodes a novel ubiquitin protein ligase that is disrupted in a18H mice. Nature Genet. 18, 143–146 (1998).
Yui, D. et al. Interchangeable binding of Bcl10 to TRAF2 and cIAPs regulates apoptosis signaling. Oncogene 20, 4317–4323 (2001).
Liu, Y. C., Penninger, J. & Karin, M. Immunity by ubiquitylation: a reversible process of modification. Nature Rev. Immunol. 5, 941–952 (2005).
Banner, D. W. et al. Crystal structure of the soluble human 55-kd TNF receptor–human TNFβ complex: implications for TNF receptor activation. Cell 73, 431–445 (1993).
Schmidt-Supprian, M. et al. Mature T cells depend on signaling through the IKK complex. Immunity 19, 377–389 (2003).
Alizadeh, A. A. et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 403, 503–511 (2000).
Hans, C. P. et al. Expression of PKCβ or cyclin D2 predicts for inferior survival in diffuse large B-cell lymphoma. Mod. Pathol. 18, 1377–1384 (2005).
Nakamura, S. et al. Overexpression of caspase recruitment domain (CARD) membrane-associated guanylate kinase 1 (CARMA1) and CARD9 in primary gastric B-cell lymphoma. Cancer 104, 1885–1893 (2005).
Ngo, V. N. et al. A loss-of-function RNA interference screen for molecular targets in cancer. Nature 441, 106–110 (2006). These authors use a library of siRNA vectors to show that CARMA1 is a major upstream signalling molecule responsible for constitutive IKK activity, which is a hallmark of B-cell lymphomas of poor prognosis.
Bertoni, F. & Zucca, E. Delving deeper into MALT lymphoma biology. J. Clin. Invest. 116, 22–26 (2006).
Billadeau, D. D. & Leibson, P. J. ITAMs versus ITIMs: striking a balance during cell regulation. J. Clin. Invest. 109, 161–168 (2002).
Lee, K. Y., D'Acquisto, F., Hayden, M. S., Shim, J. H. & Ghosh, S. PDK1 nucleates T-cell receptor-induced signaling complex for NF-κB activation. Science 308, 114–118 (2005).
Newton, A. C. Regulation of the ABC kinases by phosphorylation: protein kinase C as a paradigm. Biochem. J. 370, 361–371 (2003).
Kane, L. P. & Weiss, A. The PI3-kinase/Akt pathway and T-cell activation: pleiotropic pathways downstream of PIP3 . Immunol. Rev. 192, 7–20 (2003).
Fruman, D. A. Phosphoinositide 3-kinase and its targets in B-cell and T-cell signaling. Curr. Opin. Immunol. 16, 314–320 (2004).
Narayan, P., Holt, B., Tosti, R. & Kane, L. P. CARMA1 is required for Akt-mediated NF-κB activation in T cells. Mol. Cell Biol. 26, 2327–2336 (2006).
Bidere, N., Snow, A. L., Sakai, K., Zheng, L. & Lenardo, M. J. Caspase-8 regulation by direct interaction with TRAF6 in T-cell receptor-induced NF-κB activation. Curr. Biol. 16, 1666–1671 (2006).
Su, H. et al. Requirement for caspase-8 in NF-κB activation by antigen receptor. Science 307, 1465–1468 (2005).
Beisner, D. R., Ch'en, I. L., Kolla, R. V., Hoffmann, A. & Hedrick, S. M. Cutting edge: innate immunity conferred by B cells is regulated by caspase-8. J. Immunol. 175, 3469–3473 (2005).
Kobayashi, K. et al. RICK/Rip2/CARDIAK mediates signalling for receptors of the innate and adaptive immune systems. Nature 416, 194–199 (2002).
Chin, A. I. et al. Involvement of receptor-interacting protein 2 in innate and adaptive immune responses. Nature 416, 190–194 (2002).
Gao, M. et al. Jun turnover is controlled through JNK-dependent phosphorylation of the E3 ligase Itch. Science 306, 271–275 (2004).
Chen, Z. J. Ubiquitin signalling in the NF-κB pathway. Nature Cell Biol. 7, 758–765 (2005).
Brummelkamp, T. R., Nijman, S. M., Dirac, A. M. & Bernards, R. Loss of the cylindromatosis tumour suppressor inhibits apoptosis by activating NF-κB. Nature 424, 797–801 (2003).
Kovalenko, A. et al. The tumour suppressor CYLD negatively regulates NF-κB signalling by deubiquitination. Nature 424, 801–805 (2003).
Trompouki, E. et al. CYLD is a deubiquitinating enzyme that negatively regulates NF-κB activation by TNFR family members. Nature 424, 793–796 (2003).
Wertz, I. E. et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-κB signalling. Nature 430, 694–699 (2004).
Boone, D. L. et al. The ubiquitin-modifying enzyme A20 is required for termination of Toll-like receptor responses. Nature Immunol. 5, 1052–1060 (2004).
Acknowledgements
We thank A. Hui for assistance with the mauscript. D.J.R. is the recipient of a McDonnell Scholar Award and a Leukemia and Lymphoma Society Scholar Award. This work was supported, in part, by NIH grants and by the American Cancer Society.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Glossary
- Non-canonical pathway of NF-κB signalling
-
An alternative pathway by which the nuclear factor-κB (NF-κB) transcription factor can be released from the cytoplasm into the nucleus. Unlike NF-κB1 (p105/p50) in the classical (canonical) NF-κB pathway, NF-κB2 (p100/p52) is not pre-cleaved. An IκB (inhibitor of NF-κB) domain in the carboxyl terminus of p100 tethers it in the cytosol. Activation of specific receptors (for example, CD40) induces p100 phosphorylation by IκB kinase-α, which targets p100 for proteasomal degradation. Only the IκB domain is degraded by the proteasome, leaving the portion of p100 with transcription-factor activity (p52) intact and able to translocate to the nucleus.
- Ubiquitylation
-
The attachment of the small protein ubiquitin to lysine residues present in other proteins. Subsequent ubiquitylation events can extend from the initial ubiquitin at one of its seven lysine residues (K6, K11, K27, K29, K33, K48 or K63), forming a polyubiquitin chain.
- Lipid rafts
-
Semi-rigid lipid microdomains of the cell membrane that are enriched for sphingolipids and cholesterol. Certain signalling molecules selectively localize to lipid rafts, either constitutively or inducibly in response to a stimulus.
- E3 ubiquitin ligase
-
Protein ubiquitylation occurs in three enzymatic steps, requiring a ubiquitin-activating enzyme (E1), a ubiquitin-conjugating enzyme (E2) and a ubiquitin ligase (E3), which catalyses the ligation of an isopeptide bond between the carboxy-terminal domain of ubiquitin and an amino group belonging to a lysine residue of the target protein.
- K63-linked ubiquitylation
-
The addition of ubiquitin to lysine side chains of target proteins using the lysine at position 63 (K63) of ubiquitin. Unlike K48-linked ubiquitin chains, which are the main signal for targeting substrates for proteasomal degradation, K63-linked ubiquitin modification regulates protein function, targets certain proteins for endocytosis, and interacts with proteins with specific ubiquitin-binding domains.
- Cre–loxP system
-
A site-specific recombination system. Two short DNA sequences (loxP sites) are engineered to flank the target DNA. Expression of Cre recombinase leads to excision of the intervening sequence. Depending on the type of promoter, Cre can be expressed at specific times during development or in specific sets of cells.
Rights and permissions
About this article
Cite this article
Rawlings, D., Sommer, K. & Moreno-García, M. The CARMA1 signalosome links the signalling machinery of adaptive and innate immunity in lymphocytes. Nat Rev Immunol 6, 799–812 (2006). https://doi.org/10.1038/nri1944
Issue Date:
DOI: https://doi.org/10.1038/nri1944
This article is cited by
-
Functional implications of the CpG island methylation in the pathogenesis of celiac disease
Molecular Biology Reports (2022)
-
Fusion transcripts FYN-TRAF3IP2 and KHDRBS1-LCK hijack T cell receptor signaling in peripheral T-cell lymphoma, not otherwise specified
Nature Communications (2021)
-
Oncogenic CARMA1 couples NF-κB and β-catenin signaling in diffuse large B-cell lymphomas
Oncogene (2016)
-
The versatile roles of CARDs in regulating apoptosis, inflammation, and NF-κB signaling
Apoptosis (2015)
-
PSGR promotes prostatic intraepithelial neoplasia and prostate cancer xenograft growth through NF-κB
Oncogenesis (2014)