Key Points
-
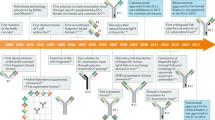
This Review discusses the theory and practices behind the generation of combinatorial antibody libraries.
-
Such libraries give scientists unprecedented control over the antibody repertoire.
-
They have led to many antibody drugs, including adalimumab (Humira; AbbVie), which is the best-selling drug in the world.
-
As combinatorial antibody libraries allow a detailed analysis of the output of the immune response, they have allowed us to understand how the adaptive immune system copes with infection when there is not sufficient time to develop a mature antibody response. This has been termed an 'SOS response'.
-
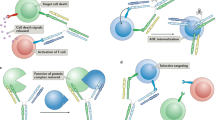
In their intracellular format, antibodies can be used as selectable proteins that perturb cell fates. Such antibodies are usually gain-of-function agonists.
-
We have moved from the engineering of antibodies to engineering with antibodies — this represents a whole new dimension in immunochemistry.
Abstract
Immunochemists have become quite proficient in engineering existing antibody molecules to control their pharmacological properties. However, in terms of generating new antibodies, the combinatorial antibody library has become a central feature of modern immunochemistry. These libraries are essentially an immune system in a test tube and enable the selection of antibodies without the constraints of whole animal or cell-based systems. This Review provides an overview of how antibody libraries are constructed and discusses what can be learnt from these synthetic systems. In particular, the Review focuses on new biological insights from antibody libraries — such as the concept of 'SOS antibodies' — and the growing use of intracellular antibodies to perturb cellular functions.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Edelman, G. M. Dissociation of γ-globulin. J. Am. Chem. Soc. 81, 3155–3156 (1959).
Porter, R. R. The hydrolysis of rabbit γ-globulin and antibodies with crystalline papain. Biochem. J. 73, 119–126 (1959).
Jones, P. T., Dear, P. H., Foote, J., Neuberger, M. S. & Winter, G. Replacing the complementarity-determining regions in a human antibody with those from a mouse. Nature 321, 522–525 (1986).
Romain, G. et al. Antibody Fc engineering improves frequency and promotes kinetic boosting of serial killing mediated by NK cells. Blood 124, 3241–3249 (2014).
Chames, P. & Baty, D. Bispecific antibodies for cancer therapy: the light at the end of the tunnel? mAbs 1, 539–547 (2009).
Hess, C., Venetz, D. & Neri, D. Emerging classes of armed antibody therapeutics against cancer. Med. Chem. Commun. 5, 408–431 (2014).
Peng, Y. J. et al. A general method for insertion of functional proteins within proteins via combinatorial selection of permissive junctions. Chem. Biol. 22, 1134–1143 (2015).
Zhang, Y. et al. Functional antibody CDR3 fusion proteins with enhanced pharmacological properties. Angew. Chem. Int. Ed. Engl. 52, 8295–8298 (2013).
Zhang, Y. et al. An antibody CDR3-erythropoietin fusion protein. ACS Chem. Biol. 8, 2117–2121 (2013).
Zhang, Y., Liu, Y., Wang, Y., Schultz, P. G. & Wang, F. Rational design of humanized dual-agonist antibodies. J. Am. Chem. Soc. 137, 38–41 (2015).
Bashford-Rogers, R. J. M. et al. Network properties derived from deep sequencing of human B-cell receptor repertoires delineate B-cell populations. Genome Res. 23, 1874–1884 (2013).
Georgiou, G. et al. The promise and challenge of high-throughput sequencing of the antibody repertoire. Nat. Biotechnol. 32, 158–168 (2014).
Lerner, R. A. Manufacturing immunity to disease in a test tube: the magic bullet realized. Angew. Chem. Int. Ed. Engl. 45, 8106–8125 (2006).
Sastry, L. et al. Cloning of the immunological repertoire in Escherichia coli for generation of monoclonal catalytic antibodies: construction of a heavy-chain variable region-specific cDNA library. Proc. Natl Acad. Sci. USA 86, 5728–5732 (1989).
Orlandi, R., Gussow, D. H., Jones, P. T. & Winter, G. Cloning immunoglobulin variable domains for expression by the polymerase chain reaction. Proc. Natl Acad. Sci. USA 86, 3833–3837 (1989).
Huse, W. D. et al. Generation of a large combinatorial library of the immunoglobulin repertoire in phage lambda. Science 246, 1275–1281 (1989). This is the original paper describing combinatorial libraries.
Mccafferty, J., Griffiths, A. D., Winter, G. & Chiswell, D. J. Phage antibodies: filamentous phage displaying antibody variable domains. Nature 348, 552–554 (1990). This study shows how antibodies can be expressed in a filamentous phage.
Barbas, C. F., Kang, A. S., Lerner, R. A. & Benkovic, S. J. Assembly of combinatorial antibody libraries on phage surfaces: the gene III site. Proc. Natl Acad. Sci. USA 88, 7978–7982 (1991). This is an early report on the phagemid format.
Lerner, R. A., Kang, A. S., Bain, J. D., Burton, D. R. & Barbas, C. F. Antibodies without Immunization. Science 258, 1313–1314 (1992).
Persson, M. A. A. Twenty years of combinatorial antibody libraries, but how well do they mimic the immunoglobulin repertoire? Proc. Natl Acad. Sci. USA 106, 20137–20138 (2009). This paper is a thorough analysis of combinatorial library content.
Paul, W. E. Fundamental Immunology (Wolters Kluwer/Lippincott Williams & Wilkins, 2008).
Wardemann, H. et al. Predominant autoantibody production by early human B cell precursors. Science 301, 1374–1377 (2003).
Melchers, F. et al. Repertoire selection by pre-B-cell receptors and B-cell receptors, and genetic control of B-cell development from immature to mature B cells. Immunol. Rev. 175, 33–46 (2000).
Smith, G. P. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science 228, 1315–1317 (1985).
Schaffitzel, C., Hanes, J., Jermutus, L. & Pluckthun, A. Ribosome display: an in vitro method for selection and evolution of antibodies from libraries. J. Immunol. Methods 231, 119–135 (1999).
Breitling, F., Dubel, S., Seehaus, T., Klewinghaus, I. & Little, M. A surface expression vector for antibody screening. Gene 104, 147–153 (1991).
Boder, E. T. & Wittrup, K. D. Yeast surface display for directed evolution of protein expression, affinity, and stability. Methods Enzymol. 328, 430–444 (2000).
Georgiou, G. et al. Display of heterologous proteins on the surface of microorganisms: from the screening of combinatorial libraries to live recombinant vaccines. Nat. Biotechnol. 15, 29–34 (1997).
Zhou, C., Jacobsen, F. W., Cai, L., Chen, Q. & Shen, W. D. Development of a novel mammalian cell surface antibody display platform. mAbs 2, 508–518 (2010).
Philippidis, A. The top 25 best-selling drugs of 2014. Genetic Engineering & Biotechnology News http://www.genengnews.com/insight-and-intelligence/the-top-25-best-selling-drugs-of-2014/77900383/#gsaccess (2015).
Taube, R. et al. Lentivirus display: stable expression of human antibodies on the surface of human cells and virus particles. PLoS ONE 3, e3181 (2008).
Ponsel, D., Neugebauer, J., Ladetzki-Baehs, K. & Tissot, K. High Affinity, developability and functional size: the holy grail of combinatorial antibody library generation. Molecules 16, 3675–3700 (2011).
Tiller, T. et al. A fully synthetic human Fab antibody library based on fixed VH/VL framework pairings with favorable biophysical properties. mAbs 5, 445–470 (2013).
Barbas, C. F., Rosenblum, J. S. & Lerner, R. A. Direct selection of antibodies that coordinate metals from semisynthetic combinatorial libraries. Proc. Natl Acad. Sci. USA 90, 6385–6389 (1993).
Kashyap, A. K. et al. Combinatorial antibody libraries from survivors of the Turkish H5N1 avian influenza outbreak reveal virus neutralization strategies. Proc. Natl Acad. Sci. USA 105, 5986–5991 (2008).
Throsby, M. et al. Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PLoS ONE 3, e3942 (2008).
Sui, J. H. et al. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat. Struct. Mol. Biol. 16, 65–273 (2009).
Lerner, R. A. Rare antibodies from combinatorial libraries suggests an S.O.S. component of the human immunological repertoire. Mol. Biosyst. 7, 1004–1012 (2011). This is the first suggestion of an SOS component of the antibody repertoire.
Ekiert, D. C. et al. Antibody recognition of a highly conserved influenza virus epitope. Science 324, 246–251 (2009).
Dreyfus, C. et al. Highly conserved protective epitopes on influenza B viruses. Science 337, 1343–1348 (2012).
Skehel, J. J. & Wiley, D. C. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 69, 531–569 (2000).
Cohn, M. & Langman, R. E. The protection: the unit of humoral immunity selected by evolution. Immunol. Rev. 115, 7–147 (1990).
Janeway, C. A. The immune system evolved to discriminate infectious nonself from noninfectious self. Immunol. Today 13, 11–16 (1992).
Lingwood, D. et al. Structural and genetic basis for development of broadly neutralizing influenza antibodies. Nature 489, 566–570 (2012).
Pappas, L. et al. Rapid development of broadly influenza neutralizing antibodies through redundant mutations. Nature 516, 418–422 (2014).
Yea, K. et al. Agonist antibody that induces human malignant cells to kill one another. Proc. Natl Acad. Sci. USA 112, E6158–E6165 (2015). This is a new approach to cancer therapy.
Xie, J., Zhang, H. K., Yea, K. & Lerner, R. A. Autocrine signaling based selection of combinatorial antibodies that transdifferentiate human stem cells. Proc. Natl Acad. Sci. USA 110, 8099–8104 (2013). This paper is a description of the power of autocrine selection.
Xie, J. et al. Prevention of cell death by antibodies selected from intracellular combinatorial libraries. Chem. Biol. 21, 274–283 (2014).
Yea, K. et al. Converting stem cells to dendritic cells by agonist antibodies from unbiased morphogenic selections. Proc. Natl Acad. Sci. USA 110, 14966–14971 (2013).
Yea, K., Xie, J., Zhang, H., Zhang, W. & Lerner, R. A. Selection of multiple agonist antibodies from intracellular combinatorial libraries reveals that cellular receptors are functionally pleiotropic. Curr. Opin. Chem. Biol. 26, 1–7 (2015). This paper is a detailed analysis of receptor pleiotropism.
Zhang, H. K., Wilson, I. A. & Lerner, R. A. Selection of antibodies that regulate phenotype from intracellular combinatorial antibody libraries. Proc. Natl Acad. Sci. USA 109, 15728–15733 (2012). This is the first description of using intracellular antibodies to regulate cell fates.
Zhang, H. K. et al. Selecting agonists from single cells infected with combinatorial antibody libraries. Chem. Biol. 20, 734–741 (2013).
Lerner, R. A. et al. Antibodies from combinatorial libraries use functional receptor pleiotropism to regulate cell fates. Q. Rev. Biophys. 48, 389–394 (2015).
Melidoni, A. N., Dyson, M. R., Wormald, S. & McCafferty, J. Selecting antagonistic antibodies that control differentiation through inducible expression in embryonic stem cells. Proc. Natl Acad. Sci. USA 110, 17802–17807 (2013).
Zebedee, S. L., et al. Human combinatorial antibody libraries to hepatitis B surface antigen. Proc. Natl Acad. Sci. USA 89, 3175–3179 (1992).
Acknowledgements
The author thanks I. Wilson, M. Pique, J. Joyce, D. Burton, R. Stanfield and W. Marisco for helpful discussions. M. Pique and I. Wilson helped with some of the figures. I thank the JPB Foundation and Zebra Biologics for their generous support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
R.A.L. has a financial interest in Zebra Biologics.
Rights and permissions
About this article
Cite this article
Lerner, R. Combinatorial antibody libraries: new advances, new immunological insights. Nat Rev Immunol 16, 498–508 (2016). https://doi.org/10.1038/nri.2016.67
Published:
Issue Date:
DOI: https://doi.org/10.1038/nri.2016.67
This article is cited by
-
Selection of a picomolar antibody that targets CXCR2-mediated neutrophil activation and alleviates EAE symptoms
Nature Communications (2021)
-
Development of therapeutic antibodies for the treatment of diseases
Journal of Biomedical Science (2020)
-
Antibody responses to viral infections: a structural perspective across three different enveloped viruses
Nature Microbiology (2019)
-
Phage Display Libraries: From Binders to Targeted Drug Delivery and Human Therapeutics
Molecular Biotechnology (2019)
-
Prospects of immunotherapy for cancer
Frontiers of Medicine (2019)