Key Points
-
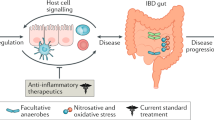
Alterations in intestinal microbial composition have long been associated with chronic inflammation; however, a definitive cause–effect relationship between dysbiosis and IBD has been difficult to prove, especially in humans
-
Dysbiosis alters not only the composition of the intestinal microbiota, but also its metabolome, thereby exerting a wide range of effects on the host
-
While the microbiota plays a key pathogenic role in IBD, chronic inflammation, in turn, promotes dysbiosis by altering the oxidative and metabolic environment of the gut
-
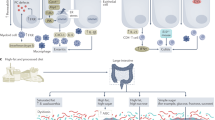
Animal studies have elucidated key immunological pathways in the pathogenesis of IBD, established both pro-inflammatory and anti-inflammatory roles of the gut microbiota, and shown that the gut microbiota is indispensable for pathogenesis in most colitis models
-
Microbial-based treatments will likely have a role in the future management of IBD; however, many questions remain regarding the bacterial composition, timing of administration, and patient selection for such therapies
Abstract
A general consensus exists that IBD is associated with compositional and metabolic changes in the intestinal microbiota (dysbiosis). However, a direct causal relationship between dysbiosis and IBD has not been definitively established in humans. Findings from animal models have revealed diverse and context-specific roles of the gut microbiota in health and disease, ranging from protective to pro-inflammatory actions. Moreover, evidence from these experimental models suggest that although gut bacteria often drive immune activation, chronic inflammation in turn shapes the gut microbiota and contributes to dysbiosis. The purpose of this Review is to summarize current associations between IBD and dysbiosis, describe the role of the gut microbiota in the context of specific animal models of colitis, and discuss the potential role of microbiota-focused interventions in the treatment of human IBD. Ultimately, more studies will be needed to define host–microbial relationships relevant to human disease and amenable to therapeutic interventions.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Dahlhamer, J. M., Zammitti, E. P., Ward, B. W., Wheaton, A. G. & Croft, J. B. Prevalence of inflammatory bowel disease among adults aged ≥18 Years — United States, 2015. MMWR Morb. Mortal. Wkly Rep. 65, 1166–1169 (2016).
Molodecky, N. A. et al. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology 142, 46–54.e42 (2012).
Manichanh, C., Borruel, N., Casellas, F. & Guarner, F. The gut microbiota in IBD. Nat. Rev. Gastroenterol. Hepatol. 9, 599–608 (2012).
Sartor, R. B. & Mazmanian, S. K. Intestinal microbes in inflammatory bowel diseases. Am. J. Gastroenterol. Suppl. 1, 15–21 (2012).
Mosca, A., Leclerc, M. & Hugot, J. P. Gut microbiota diversity and human diseases: should we reintroduce key predators in our ecosystem? Front. Microbiol. 7, 455 (2016).
Ott, S. J. et al. Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut 53, 685–693 (2004).
Manichanh, C. et al. Reduced diversity of faecal microbiota in Crohn's disease revealed by a metagenomic approach. Gut 55, 205–211 (2006).
Khan, K. J. et al. Antibiotic therapy in inflammatory bowel disease: a systematic review and meta-analysis. Am. J. Gastroenterol. 106, 661–673 (2011).
Wang, S. L., Wang, Z. R. & Yang, C. Q. Meta-analysis of broad-spectrum antibiotic therapy in patients with active inflammatory bowel disease. Exp. Ther. Med. 4, 1051–1056 (2012).
Harper, P. H., Lee, E. C., Kettlewell, M. G., Bennett, M. K. & Jewell, D. P. Role of the faecal stream in the maintenance of Crohn's colitis. Gut 26, 279–284 (1985).
Rutgeerts, P. et al. Effect of faecal stream diversion on recurrence of Crohn's disease in the neoterminal ileum. Lancet 338, 771–774 (1991).
Janowitz, H. D., Croen, E. C. & Sachar, D. B. The role of the fecal stream in Crohn's disease: an historical and analytic review. Inflamm. Bowel Dis. 4, 29–39 (1998).
Kaplan, G. G. & Ng, S. C. Understanding and preventing the global increase of inflammatory bowel disease. Gastroenterology 152, 313–321.e2 (2017).
De Filippo, C. et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl Acad. Sci. USA 107, 14691–14696 (2010).
Sokol, H. et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl Acad. Sci. USA 105, 16731–16736 (2008).
Van de Merwe, J. P., Schroder, A. M., Wensinck, F. & Hazenberg, M. P. The obligate anaerobic faecal flora of patients with Crohn's disease and their first-degree relatives. Scand. J. Gastroenterol. 23, 1125–1131 (1988).
Walker, A. W. et al. High-throughput clone library analysis of the mucosa-associated microbiota reveals dysbiosis and differences between inflamed and non-inflamed regions of the intestine in inflammatory bowel disease. BMC Microbiol. 11, 7 (2011).
Gophna, U., Sommerfeld, K., Gophna, S., Doolittle, W. F. & Veldhuyzen van Zanten, S. J. Differences between tissue-associated intestinal microfloras of patients with Crohn's disease and ulcerative colitis. J. Clin. Microbiol. 44, 4136–4141 (2006).
Frank, D. N. et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl Acad. Sci. USA 104, 13780–13785 (2007). Culture-independent phylogenetic analysis to characterize the gut microbiotas of patients with IBD and non-IBD controls showed that a subset of Crohn's disease and ulcerative colitis samples contained abnormal gut microbiota, characterized by depletion of commensal bacteria, notably members of the phyla Firmicutes and Bacteroidetes.
Martinez-Medina, M., Aldeguer, X., Gonzalez-Huix, F., Acero, D. & Garcia-Gil, L. J. Abnormal microbiota composition in the ileocolonic mucosa of Crohn's disease patients as revealed by polymerase chain reaction-denaturing gradient gel electrophoresis. Inflamm. Bowel Dis. 12, 1136–1145 (2006).
Prescott, N. J. et al. A nonsynonymous SNP in ATG16L1 predisposes to ileal Crohn's disease and is independent of CARD15 and IBD5. Gastroenterology 132, 1665–1671 (2007).
Swidsinski, A., Loening-Baucke, V., Vaneechoutte, M. & Doerffel, Y. Active Crohn's disease and ulcerative colitis can be specifically diagnosed and monitored based on the biostructure of the fecal flora. Inflamm. Bowel Dis. 14, 147–161 (2008).
Rehman, A. et al. Geographical patterns of the standing and active human gut microbiome in health and IBD. Gut 65, 238–248 (2016).
Halfvarson, J. et al. Dynamics of the human gut microbiome in inflammatory bowel disease. Nat. Microbiol. 2, 17004 (2017).
Hansen, R. et al. Microbiota of de-novo pediatric IBD: increased Faecalibacterium prausnitzii and reduced bacterial diversity in Crohn's but not in ulcerative colitis. Am. J. Gastroenterol. 107, 1913–1922 (2012).
Assa, A. et al. Mucosa-associated ileal microbiota in new-onset pediatric crohn's disease. Inflamm. Bowel Dis. 22, 1533–1539 (2016).
Seksik, P. et al. Alterations of the dominant faecal bacterial groups in patients with Crohn's disease of the colon. Gut 52, 237–242 (2003).
Baumgart, M. et al. Culture independent analysis of ileal mucosa reveals a selective increase in invasive Escherichia coli of novel phylogeny relative to depletion of Clostridiales in Crohn's disease involving the ileum. ISME J. 1, 403–418 (2007).
Sartor, R. B. Therapeutic correction of bacterial dysbiosis discovered by molecular techniques. Proc. Natl Acad. Sci. USA 105, 16413–16414 (2008).
Mangin, I. et al. Molecular inventory of faecal microflora in patients with Crohn's disease. FEMS Microbiol. Ecol. 50, 25–36 (2004).
Joossens, M. et al. Dysbiosis of the faecal microbiota in patients with Crohn's disease and their unaffected relatives. Gut 60, 631–637 (2011).
Willing, B. P. et al. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology 139, 1844–1854.e1 (2010).
Lepage, P. et al. Twin study indicates loss of interaction between microbiota and mucosa of patients with ulcerative colitis. Gastroenterology 141, 227–236 (2011).
Shah, R. et al. Composition and function of the pediatric colonic mucosal microbiome in untreated patients with ulcerative colitis. Gut Microbes 7, 384–396 (2016).
Kellermayer, R. et al. Microbiota separation and C-reactive protein elevation in treatment-naive pediatric granulomatous Crohn disease. J. Pediatr. Gastroenterol. Nutr. 55, 243–250 (2012).
Gevers, D. et al. The treatment-naive microbiome in new-onset Crohn's disease. Cell Host Microbe 15, 382–392 (2014). This paper analysed microbiomes from multiple gastrointestinal locations in new-onset Crohn's disease cases, identifying co-occurring and co-excluded Crohn's-disease-associated microorganism and finding that antibiotics amplify the dysbiosis associated with Crohn's disease.
Lewis, J. D. et al. Inflammation, antibiotics, and diet as environmental stressors of the gut microbiome in pediatric Crohn's disease. Cell Host Microbe 18, 489–500 (2015).
Chow, J., Tang, H. & Mazmanian, S. K. Pathobionts of the gastrointestinal microbiota and inflammatory disease. Curr. Opin. Immunol. 23, 473–480 (2011).
Darfeuille-Michaud, A. et al. Presence of adherent Escherichia coli strains in ileal mucosa of patients with Crohn's disease. Gastroenterology 115, 1405–1413 (1998).
Boudeau, J., Glasser, A. L., Masseret, E., Joly, B. & Darfeuille-Michaud, A. Invasive ability of an Escherichia coli strain isolated from the ileal mucosa of a patient with Crohn's disease. Infect. Immun. 67, 4499–4509 (1999).
Chiodini, R. J., Van Kruiningen, H. J., Thayer, W. R., Merkal, R. S. & Coutu, J. A. Possible role of mycobacteria in inflammatory bowel disease. I. An unclassified Mycobacterium species isolated from patients with Crohn's disease. Dig. Dis. Sci. 29, 1073–1079 (1984).
Feller, M. et al. Mycobacterium avium subspecies paratuberculosis and Crohn's disease: a systematic review and meta-analysis. Lancet Infect. Dis. 7, 607–613 (2007).
Wagner, J. et al. Mycobacterium avium subspecies paratuberculosis in children with early-onset Crohn's disease: a longitudinal follow-up study. Inflamm. Bowel Dis. 17, 1825–1826 (2011).
Timms, V. J., Daskalopoulos, G., Mitchell, H. M. & Neilan, B. A. The association of Mycobacterium avium subsp. paratuberculosis with inflammatory bowel disease. PLoS ONE 11, e0148731 (2016).
Selby, W. et al. Two-year combination antibiotic therapy with clarithromycin, rifabutin, and clofazimine for Crohn's disease. Gastroenterology 132, 2313–2319 (2007).
Liverani, E., Scaioli, E., Cardamone, C., Dal Monte, P. & Belluzzi, A. Mycobacterium avium subspecies paratuberculosis in the etiology of Crohn's disease, cause or epiphenomenon? World J. Gastroenterol. 20, 13060–13070 (2014).
Dharmani, P., Strauss, J., Ambrose, C., Allen-Vercoe, E. & Chadee, K. Fusobacterium nucleatum infection of colonic cells stimulates MUC2 mucin and tumor necrosis factor alpha. Infect. Immun. 79, 2597–2607 (2011).
Zeller, G. et al. Potential of fecal microbiota for early-stage detection of colorectal cancer. Mol. Syst. Biol. 10, 766 (2014).
Glover, L. E., Lee, J. S. & Colgan, S. P. Oxygen metabolism and barrier regulation in the intestinal mucosa. J. Clin. Invest. 126, 3680–3688 (2016).
Marteyn, B. et al. Modulation of Shigella virulence in response to available oxygen in vivo. Nature 465, 355–358 (2010).
Albenberg, L. et al. Correlation between intraluminal oxygen gradient and radial partitioning of intestinal microbiota in humans and mice. Gastroenterology 147, 1055–1063.e8 (2014).
Lopez, C. A. et al. Virulence factors enhance Citrobacter rodentium expansion through aerobic respiration. Science 353, 1249–1253 (2016).
Winter, S. E., Lopez, C. A. & Baumler, A. J. The dynamics of gut-associated microbial communities during inflammation. EMBO Rep. 14, 319–327 (2013).
Breitbart, M. et al. Metagenomic analyses of an uncultured viral community from human feces. J. Bacteriol. 185, 6220–6223 (2003).
Reyes, A. et al. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature 466, 334–338 (2010).
Minot, S. et al. The human gut virome: inter-individual variation and dynamic response to diet. Genome Res. 21, 1616–1625 (2011).
Wagner, J. et al. Bacteriophages in gut samples from pediatric Crohn's disease patients: metagenomic analysis using 454 pyrosequencing. Inflamm. Bowel Dis. 19, 1598–1608 (2013).
Norman, J. M. et al. Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell 160, 447–460 (2015). This study analysed the enteric virome, which was abnormal in patients with Crohn's disease and ulcerative colitis and had a substantial expansion of Caudovirales bacteriophages.
Magin, W. S., Van Kruiningen, H. J. & Colombel, J. F. Immunohistochemical search for viral and bacterial antigens in Crohn's disease. J. Crohns Colitis 7, 161–166 (2013).
Main, J. et al. Antibody to Saccharomyces cerevisiae (bakers' yeast) in Crohn's disease. BMJ 297, 1105–1106 (1988).
Nilsson, R. H. et al. Taxonomic reliability of DNA sequences in public sequence databases: a fungal perspective. PLoS ONE 1, e59 (2006).
Underhill, D. M. & Iliev, I. D. The mycobiota: interactions between commensal fungi and the host immune system. Nat. Rev. Immunol. 14, 405–416 (2014).
Findley, K. et al. Topographic diversity of fungal and bacterial communities in human skin. Nature 498, 367–370 (2013).
Sokol, H. et al. Fungal microbiota dysbiosis in IBD. Gut 66, 1039–1048 (2016).
Ott, S. J. et al. Fungi and inflammatory bowel diseases: alterations of composition and diversity. Scand. J. Gastroenterol. 43, 831–841 (2008).
Mukhopadhya, I. et al. The fungal microbiota of de-novo paediatric inflammatory bowel disease. Microbes Infect. 17, 304–310 (2015).
Liguori, G. et al. Fungal dysbiosis in mucosa-associated microbiota of Crohn's disease patients. J. Crohns Colitis 10, 296–305 (2016).
Richard, M. L., Lamas, B., Liguori, G., Hoffmann, T. W. & Sokol, H. Gut fungal microbiota: the Yin and Yang of inflammatory bowel disease. Inflamm. Bowel Dis. 21, 656–665 (2015).
Iliev, I. D. et al. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science 336, 1314–1317 (2012). This study shows that mice lacking dectin-1 exhibited increased susceptibility to chemically induced colitis, which was the result of altered responses to indigenous fungi; in humans, a polymorphism in the gene for dectin-1 ( CLEC7A ) was strongly linked to a severe form of ulcerative colitis.
Lamas, B. et al. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat. Med. 22, 598–605 (2016).
Chiaro, T. R. et al. A member of the gut mycobiota modulates host purine metabolism exacerbating colitis in mice. Sci. Transl Med. 9, eaaf9044 (2017).
Wheeler, M. L. et al. Immunological consequences of intestinal fungal dysbiosis. Cell Host Microbe 19, 865–873 (2016).
Wishart, D. S. et al. HMDB: the Human Metabolome Database. Nucleic Acids Res. 35, D521–D526 (2007).
Morgan, X. C. et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 13, R79 (2012).
Gadaleta, R. M. et al. Farnesoid X receptor activation inhibits inflammation and preserves the intestinal barrier in inflammatory bowel disease. Gut 60, 463–472 (2011). Nuclear farnesoid X receptor is a bile salt that is implicated in intestinal antibacterial defence and barrier function; this study shows that its activation prevents chemically induced intestinal inflammation in mice and inhibits pro-inflammatory cytokine production.
Ogilvie, L. A. & Jones, B. V. Dysbiosis modulates capacity for bile acid modification in the gut microbiomes of patients with inflammatory bowel disease: a mechanism and marker of disease? Gut 61, 1642–1643 (2012).
Duboc, H. et al. Connecting dysbiosis, bile-acid dysmetabolism and gut inflammation in inflammatory bowel diseases. Gut 62, 531–539 (2013).
Smith, P. M. et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 341, 569–573 (2013).
Atarashi, K. et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science 331, 337–341 (2011).
Grigg, J. B. & Sonnenberg, G. F. Host–microbiota interactions shape local and systemic inflammatory diseases. J. Immunol. 198, 564–571 (2017).
Omenetti, S. & Pizarro, T. T. The Treg/Th17 axis: a dynamic balance regulated by the gut microbiome. Front. Immunol. 6, 639 (2015).
Rakoff-Nahoum, S., Paglino, J., Eslami-Varzaneh, F., Edberg, S. & Medzhitov, R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell 118, 229–241 (2004).
Abt, M. C. et al. Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity 37, 158–170 (2012).
Shaw, S. Y., Blanchard, J. F. & Bernstein, C. N. Association between the use of antibiotics in the first year of life and pediatric inflammatory bowel disease. Am. J. Gastroenterol. 105, 2687–2692 (2010).
van Nood, E. et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N. Engl. J. Med. 368, 407–415 (2013).
Rivas, M. A. et al. Deep resequencing of GWAS loci identifies independent rare variants associated with inflammatory bowel disease. Nat. Genet. 43, 1066–1073 (2011).
Barrett, J. C. et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat. Genet. 40, 955–962 (2008).
Sandborn, W. et al. O-001 A multicenter, double-blind, placebo-controlled phase3 study of ustekinumab, a human IL-12/23P40 mAB, in moderate-service Crohn's disease refractory to anti-TNFalpha: UNITI-1 [abstract]. Inflamm. Bowel Dis. 22 (Suppl. 1), S1 (2016).
Almeida, F. F. & Belz, G. T. Innate lymphoid cells: models of plasticity for immune homeostasis and rapid responsiveness in protection. Mucosal Immunol. 9, 1103–1112 (2016).
Ignacio, A., Morales, C. I., Camara, N. O. & Almeida, R. R. Innate sensing of the gut microbiota: modulation of inflammatory and autoimmune diseases. Front. Immunol. 7, 54 (2016).
Mohamed, R. & Lord, G. M. T-Bet as a key regulator of mucosal immunity. Immunology 147, 367–376 (2016).
Kiesler, P., Fuss, I. J. & Strober, W. Experimental models of inflammatory bowel diseases. Cell. Mol. Gastroenterol. Hepatol. 1, 154–170 (2015).
Globig, A. M. et al. Comprehensive intestinal T helper cell profiling reveals specific accumulation of IFN-gamma+IL-17+ coproducing CD4+ T cells in active inflammatory bowel disease. Inflamm. Bowel Dis. 20, 2321–2329 (2014).
Ramesh, R. et al. Pro-inflammatory human Th17 cells selectively express P-glycoprotein and are refractory to glucocorticoids. J. Exp. Med. 211, 89–104 (2014).
Stepankova, R. et al. Segmented filamentous bacteria in a defined bacterial cocktail induce intestinal inflammation in SCID mice reconstituted with CD45RBhigh CD4+ T cells. Inflamm. Bowel Dis. 13, 1202–1211 (2007).
Jiang, H. Q., Kushnir, N., Thurnheer, M. C., Bos, N. A. & Cebra, J. J. Monoassociation of SCID mice with Helicobacter muridarum, but not four other enterics, provokes IBD upon receipt of T cells. Gastroenterology 122, 1346–1354 (2002).
Cahill, R. J. et al. Inflammatory bowel disease: an immunity-mediated condition triggered by bacterial infection with Helicobacter hepaticus. Infect. Immun. 65, 3126–3131 (1997).
Mazmanian, S. K., Round, J. L. & Kasper, D. L. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 453, 620–625 (2008).
Round, J. L. & Mazmanian, S. K. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc. Natl Acad. Sci. USA 107, 12204–12209 (2010). This study showed that Bacteroides fragilis capsular polysaccharide A (PSA) can alleviate inflammation in several colitis mouse models by promoting development of regulatory T cells.
Garrett, W. S. et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe 8, 292–300 (2010).
Garrett, W. S. et al. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell 131, 33–45 (2007). This study shows that T-bet deficiency in the innate immune system results in spontaneous and communicable ulcerative colitis, and T-bet−/−RAG−/− ulcerative colitis (TRUC) mice can transmit colitis even to wild-type animals.
Veldhoen, M. et al. The aryl hydrocarbon receptor links TH17-cell-mediated autoimmunity to environmental toxins. Nature 453, 106–109 (2008).
Quintana, F. J. et al. Control of Treg and TH17 cell differentiation by the aryl hydrocarbon receptor. Nature 453, 65–71 (2008).
Alroqi, F. J. & Chatila, T. A. T regulatory cell biology in health and disease. Curr. Allergy Asthma Rep. 16, 27 (2016).
Fantini, M. C. et al. Smad7 controls resistance of colitogenic T cells to regulatory T cell-mediated suppression. Gastroenterology 136, 1308–1316.e1–e3 (2009).
Monteleone, G. et al. Mongersen, an oral SMAD7 antisense oligonucleotide, and Crohn's disease. N. Engl. J. Med. 372, 1104–1113 (2015).
Franke, A. et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nat. Genet. 42, 1118–1125 (2010).
Franke, A. et al. Sequence variants in IL10. ARPC2 and multiple other loci contribute to ulcerative colitis susceptibility. Nat. Genet. 40, 1319–1323 (2008).
Hoshi, N. et al. MyD88 signalling in colonic mononuclear phagocytes drives colitis in IL-10-deficient mice. Nat. Commun. 3, 1120 (2012).
Rakoff-Nahoum, S., Hao, L. & Medzhitov, R. Role of Toll-like receptors in spontaneous commensal-dependent colitis. Immunity 25, 319–329 (2006). This study demonstrated a differential role of innate immune recognition by Toll-like receptors (TLRs) in the development of commensal-dependent colitis and a key pathogenic role of TLR signalling for chronic intestinal inflammation.
Atarashi, K. et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 500, 232–236 (2013).
Asquith, M. & Powrie, F. An innately dangerous balancing act: intestinal homeostasis, inflammation, and colitis-associated cancer. J. Exp. Med. 207, 1573–1577 (2010).
De Jager, P. L. et al. The role of the Toll receptor pathway in susceptibility to inflammatory bowel disease. Genes Immun. 8, 387–397 (2007).
Hugot, J. P. et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature 411, 599–603 (2001).
Ogura, Y. et al. A frameshit mutation in NOD2 associated with susceptibility to Crohn's disease. Nature 411, 603–606 (2001).
Pierek, M. et al. Toll-like receptor-1, -2. and -6 polymorphisms influence disease extension in inflammatory bowel diseases. Inflamm. Bowel Dis. 12, 1–8 (2006).
Torok, H. P. et al. Crohn's disease is associated with Toll-like receptor-9 polymorphism. Gastroenterology 127, 365–366 (2004).
Villani, A. C. et al. Common variants in the NLRP3 region contribute to Crohn's disease susceptibility. Nat. Genet. 41, 71–76 (2009).
Brown, S. L. et al. Myd88-dependent positioning of Ptgs2-expressing stromal cells maintains colonic epithelial proliferation during injury. J. Clin. Invest. 117, 258–269 (2007).
Kim, J. Y., Kajino-Sakamoto, R., Omori, E., Jobin, C. & Ninomiya-Tsuji, J . Intestinal epithelial-derived TAK1 signaling is essential for cytoprotection against chemical-induced colitis. PLoS ONE 4, e4561 (2009).
Malvin, N. P., Seno, H. & Stappenbeck, T. S. . Colonic epithelial response to injury requires Myd88 signaling in myeloid cells. Mucosal Immunol. 5, 194–206 (2012).
Podolsky, D. K., Gerken, G., Eyking, A. & Cario, E. Colitis-associated variant of TLR2 causes impaired mucosal repair because of TFF3 deficiency. Gastroenterology 137, 209–220 (2009).
Pull, S. L., Doherty, J. M., Mills, J. C., Gordon, J. I. & Stappenbeck, T. S. Activated macrophages are an adaptive element of the colonic epithelial progenitor niche necessary for regenerative responses to injury. Proc. Natl Acad. Sci. USA 102, 99–104 (2005).
Zheng, L., Riehl, T. E. & Stenson, W. Regulation of colonic epithelial repair in mice by Toll-like receptors and hyaluronic acid. Gastroenterology 137, 2041–2051 (2009).
Nenci, A. et al. Epithelial NEMO links innate immunity to chronic intestinal inflammation. Nature 446, 557–561 (2007).
Allen, I. C. et al. The NLRP3 inflammasome functions as a negative regulator of tumorigenesis during colitis-associated cancer. J. Exp. Med. 207, 1045–1056 (2010).
Dupaul-Chicoine, J. et al. Control of intestinal homeostasis, colitis, and colitis-associated colorectal cancer by the inflammatory caspases. Immunity 32, 367–378 (2010).
Zaki, M. H. et al. The NLRP3 inflammasome protects against loss of epithelial integrity and mortality during experimental colitis. Immunity 32, 379–391 (2010).
Takagi, H. et al. Contrasting action of IL-12 and IL-18 in the development of dextran sodium sulphate colitis in mice. Scand. J. Gastroenterol. 38, 837–844 (2003).
Elinav, E. et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell 145, 745–757 (2011).
Gibson, D. L. et al. MyD88 signalling plays a critical role in host defence by controlling pathogen burden and promoting epithelial cell homeostasis during Citrobacter rodentium-induced colitis. Cell. Microbiol. 10, 618–631 (2008).
Lebeis, S. L., Bommarius, B., Parkos, C. A., Sherman, M. A. & Kalman, D. TLR signaling mediated by MyD88 is required for a protective innate immune response by neutrophils to Citrobacter rodentium. J. Immunol. 179, 566–577 (2007).
Cadwell, K. Crosstalk between autophagy and inflammatory signalling pathways: balancing defence and homeostasis. Nat. Rev. Immunol. 16, 661–675 (2016).
Hampe, J. et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat. Genet. 39, 207–211 (2007).
Parkes, M. et al. Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn's disease susceptibility. Nat. Genet. 39, 830–832 (2007).
Travassos, L. H. et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat. Immunol. 11, 55–62 (2010).
Cooney, R. et al. NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nat. Med. 16, 90–97 (2010).
Cadwell, K. et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature 456, 259–263 (2008).
Salzman, N. H. et al. Enteric defensins are essential regulators of intestinal microbial ecology. Nat. Immunol. 11, 76–83 (2010).
Wehkamp, J. et al. Reduced Paneth cell alpha-defensins in ileal Crohn's disease. Proc. Natl Acad. Sci. USA 102, 18129–18134 (2005).
Cadwell, K. et al. Virus-plus-susceptibility gene interaction determines Crohn's disease gene Atg16L1 phenotypes in intestine. Cell 141, 1135–1145 (2010). This study demonstrates that an interaction between a specific virus infection (murine norovirus) and a gene mutation induces Crohn's-disease-like intestinal pathologies in mice.
Schaubeck, M. et al. Dysbiotic gut microbiota causes transmissible Crohn's disease-like ileitis independent of failure in antimicrobial defence. Gut 65, 225–237 (2016).
Kontoyiannis, D., Pasparakis, M., Pizarro, T. T., Cominelli, F. & Kollias, G. Impaired on/off regulation of TNF biosynthesis in mice lacking TNF AU-rich elements: implications for joint and gut-associated immunopathologies. Immunity 10, 387–398 (1999).
Pabst, O. New concepts in the generation and functions of IgA. Nat. Rev. Immunol. 12, 821–832 (2012).
Palm, N. W. et al. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell 158, 1000–1010 (2014). This paper shows that IgA coating of intestinal bacteria can distinguish colitogenic (IgA+) from commensal (IgA−) species, and IgA+ bacteria identified in this manner include known pathobionts (Prevotellaceae, Helicobacter and segmented filamentous bacteria) capable of mediating chemically induced colitis when transferred to specific pathogen-free mice.
Ludvigsson, J. F., Neovius, M. & Hammarstrom, L. Association between IgA deficiency and other autoimmune conditions: a population-based matched cohort study. J. Clin. Immunol. 34, 444–451 (2014).
D'Haens, G. R. et al. Early lesions of recurrent Crohn's disease caused by infusion of intestinal contents in excluded ileum. Gastroenterology 114, 262–267 (1998).
Harig, J. M., Soergel, K. H., Komorowski, R. A. & Wood, C. M. Treatment of diversion colitis with short-chain-fatty acid irrigation. N. Engl. J. Med. 320, 23–28 (1989).
Sanders, M. E. et al. An update on the use and investigation of probiotics in health and disease. Gut 62, 787–796 (2013).
Singh, S., Stroud, A. M., Holubar, S. D., Sandborn, W. J. & Pardi, D. S. Treatment and prevention of pouchitis after ileal pouch-anal anastomosis for chronic ulcerative colitis. Cochrane Database Syst. Rev. 6, CD001176 (2015).
Terao, S. et al. Antibiotic combination therapy for steroid withdrawal in steroid-dependent ulcerative colitis. Digestion 83, 198–203 (2011).
Ohkusa, T. et al. Newly developed antibiotic combination therapy for ulcerative colitis: a double-blind placebo-controlled multicenter trial. Am. J. Gastroenterol. 105, 1820–1829 (2010).
Uehara, T. et al. Efficacy of antibiotic combination therapy in patients with active ulcerative colitis, including refractory or steroid-dependent cases. J. Gastroenterol. Hepatol. 25 (Suppl. 1), S62–S66 (2010).
Albenberg, L. G. & Wu, G. D. Diet and the intestinal microbiome: associations, functions, and implications for health and disease. Gastroenterology 146, 1564–1572 (2014).
Lewis, J. D. & Abreu, M. T. Diet as a trigger or therapy for inflammatory bowel diseases. Gastroenterology 152, 398–414.e6 (2017).
Sandhu, B. K. et al. Guidelines for the management of inflammatory bowel disease in children in the United Kingdom. J. Pediatr. Gastroenterol. Nutr. 50 (Suppl. 1), S1–S13 (2010).
Caprilli, R. et al. European evidence based consensus on the diagnosis and management of Crohn's disease: special situations. Gut 55 (Suppl. 1), i36–i58 (2006).
Fell, J. M. et al. Mucosal healing and a fall in mucosal pro-inflammatory cytokine mRNA induced by a specific oral polymeric diet in paediatric Crohn's disease. Aliment. Pharmacol. Ther. 14, 281–289 (2000).
Borrelli, O. et al. Polymeric diet alone versus corticosteroids in the treatment of active pediatric Crohn's disease: a randomized controlled open-label trial. Clin. Gastroenterol. Hepatol. 4, 744–753 (2006).
Afzal, N. A. et al. Improvement in quality of life of children with acute Crohn's disease does not parallel mucosal healing after treatment with exclusive enteral nutrition. Aliment. Pharmacol. Ther. 20, 167–172 (2004).
Froslie, K. F., Jahnsen, J., Moum, B. A., Vatn, M. H. & Group, I. Mucosal healing in inflammatory bowel disease: results from a Norwegian population-based cohort. Gastroenterology 133, 412–422 (2007).
Lionetti, P. et al. Enteral nutrition and microflora in pediatric Crohn's disease. JPEN J. Parenter. Enteral Nutr. 29 (4 Suppl.), S173–S175 (2005).
Leach, S. T., Mitchell, H. M., Eng, W. R., Zhang, L. & Day, A. S. Sustained modulation of intestinal bacteria by exclusive enteral nutrition used to treat children with Crohn's disease. Aliment. Pharmacol. Ther. 28, 724–733 (2008).
Tjellstrom, B. et al. Effect of exclusive enteral nutrition on gut microflora function in children with Crohn's disease. Scand. J. Gastroenterol. 47, 1454–1459 (2012).
Schwerd, T. et al. Exclusive enteral nutrition in active pediatric Crohn disease: effects on intestinal microbiota and immune regulation. J. Allergy Clin. Immunol. 138, 592–596 (2016).
Gerasimidis, K. et al. Decline in presumptively protective gut bacterial species and metabolites are paradoxically associated with disease improvement in pediatric Crohn's disease during enteral nutrition. Inflamm. Bowel Dis. 20, 861–871 (2014).
Gough, E., Shaikh, H. & Manges, A. R. Systematic review of intestinal microbiota transplantation (fecal bacteriotherapy) for recurrent Clostridium difficile infection. Clin. Infect. Dis. 53, 994–1002 (2011).
Angelberger, S. et al. Temporal bacterial community dynamics vary among ulcerative colitis patients after fecal microbiota transplantation. Am. J. Gastroenterol. 108, 1620–1630 (2013).
Cui, B. et al. Step-up fecal microbiota transplantation strategy: a pilot study for steroid-dependent ulcerative colitis. J. Transl Med. 13, 298 (2015).
Kunde, S. et al. Safety, tolerability, and clinical response after fecal transplantation in children and young adults with ulcerative colitis. J. Pediatr. Gastroenterol. Nutr. 56, 597–601 (2013).
Suskind, D. L., Singh, N., Nielson, H. & Wahbeh, G. Fecal microbial transplant via nasogastric tube for active pediatric ulcerative colitis. J. Pediatr. Gastroenterol. Nutr. 60, 27–29 (2015).
Paramsothy, S. et al. Multidonor intensive faecal microbiota transplantation for active ulcerative colitis: a randomised placebo-controlled trial. Lancet 389, 1218–1228 (2017). This randomized, placebo-controlled clinical trial of faecal microbiota transplantation (FMT) for treatment of ulcerative colitis showed that intensive-dosing, multidonor FMT induces clinical remission and endoscopic improvement in active ulcerative colitis and is associated with distinct microbial changes that relate to outcome.
Moayyedi, P. et al. Fecal microbiota transplantation induces remission in patients with active ulcerative colitis in a randomized controlled trial. Gastroenterology 149, 102–109.e6 (2015). First randomized, placebo-controlled trial of faecal microbiota transplantation in ulcerative colitis showing potential benefit.
Rossen, N. G. et al. Findings from a randomized controlled trial of fecal transplantation for patients with ulcerative colitis. Gastroenterology 149, 110–118.e4 (2015).
US National Library of Medicine. ClinicalTrials.gov [online] https://clinicaltrials.gov/ct2/show/NCT03078803, (2017).
US National Library of Medicine. ClinicalTrials.gov [online] https://clinicaltrials.gov/ct2/show/NCT02097797, (2016).
Suskind, D. L. et al. Fecal microbial transplant effect on clinical outcomes and fecal microbiome in active Crohn's disease. Inflamm. Bowel Dis. 21, 556–563 (2015).
Cui, B. et al. Fecal microbiota transplantation through mid-gut for refractory Crohn's disease: safety, feasibility, and efficacy trial results. J. Gastroenterol. Hepatol. 30, 51–58 (2015).
Kolho, K. L. et al. Fecal microbiota in pediatric inflammatory bowel disease and its relation to inflammation. Am. J. Gastroenterol. 110, 921–930 (2015).
Hildebrand, F. et al. Inflammation-associated enterotypes, host genotype, cage and inter-individual effects drive gut microbiota variation in common laboratory mice. Genome Biol. 14, R4 (2013).
Turpin, W. et al. Assoication of host genome with intestinal microbiota composition in a large healthy cohort. Nat. Genet. 48, 1413–1417 (2016).
Stein, M. M. et al. Innate immunity and asthma risk in Amish and Hutterite farm children. N. Engl. J. Med. 375, 411–421 (2016).
Olszak, T. et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science 336, 489–493 (2012).
Cho, I. et al. Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature 488, 621–626 (2012).
Acknowledgements
J.N. acknowledges support from the National Institute of Diabetes and Digestive and Kidney Diseases (T32DK007066-40) and holds the AGA-Takeda Pharmaceuticals Research Scholar Award in Inflammatory Bowel Disease. G.D.W. acknowledges funding from PennCHOP Microbiome Program, NIH grants R01 DK107565, R24 AI 118629, and the Crohn's and Colitis Foundation Microbiome Initiative. L.A. acknowledges support from grant K23DK109136-01. V.T.T. acknowledges support from NIH grant K08-DK097301.
Author information
Authors and Affiliations
Contributions
All authors contributed equally to all aspects of this manuscript.
Corresponding author
Ethics declarations
Competing interests
Seres Therapeutics has an option agreement with the University of Pennsylvania for some intellectual property, listing G.D.W. as an inventor.
Related links
FURTHER INFORMATION
PowerPoint slides
Glossary
- Crohn's disease
-
A chronic inflammatory bowel disease that can involve the entire gastrointestinal tract and is characterized by areas of transmural inflammation surrounded by normal mucosa.
- Ulcerative colitis
-
A chronic inflammatory bowel disease characterized by diffuse mucosal inflammation limited to the colon.
- Dysbiosis
-
An alteration of the microbiota that is associated with disease.
- Faecal diversion
-
Surgical diversion of the faecal stream by means of a loop ileostomy or colostomy.
- Virome
-
The collection of prokaryotic and eukaryotic viruses that are part of the human microbiota.
- Bacteriophages
-
Viruses that infect and replicate within bacteria.
- Bile acids
-
Steroid acids found in bile that aid in fat emulsification and nutrient digestion.
- Short-chain fatty acids
-
Fatty acids with less than 6 carbons produced by bacterial fermentation of dietary carbohydrates.
- Immunoglobulin A
-
(IgA). The most abundant antibody type, mostly associated with mucosal surfaces.
- Ileocolonic resection
-
Resection of the terminal ileum, caecum and ascending colon, followed by an ileocolonic anastomosis.
- Ileostomy
-
A surgical operation in which a piece of the ileum is diverted to an artificial opening in the abdominal wall.
- Probiotics
-
Living microorganisms which, when administered in adequate amounts, might confer health benefits on the host.
- Enteral nutritional therapy
-
(ENT). Nutritional supplementation via a nasoenteric feeding tube.
- Faecal microbiota transplantation
-
(FMT). The administration of microorganisms derived from the stool of a healthy donor to the gastrointestinal tract of a patient.
- Coprophagia
-
The consumption of faeces.
Rights and permissions
About this article
Cite this article
Ni, J., Wu, G., Albenberg, L. et al. Gut microbiota and IBD: causation or correlation?. Nat Rev Gastroenterol Hepatol 14, 573–584 (2017). https://doi.org/10.1038/nrgastro.2017.88
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrgastro.2017.88
This article is cited by
-
Galectin from Trichinella spiralis alleviates DSS-induced colitis in mice by regulating the intestinal microbiota
Veterinary Research (2024)
-
Transcriptome-wide association studies associated with Crohn’s disease: challenges and perspectives
Cell & Bioscience (2024)
-
Porphyromonas gingivalis aggravates colitis via a gut microbiota-linoleic acid metabolism-Th17/Treg cell balance axis
Nature Communications (2024)
-
Oral pyroptosis nanoinhibitor for the treatment of inflammatory bowel disease
Nano Research (2024)
-
Protective effects of topiramate on acetic acid-induced colitis in rats through the inhibition of oxidative stress
Naunyn-Schmiedeberg's Archives of Pharmacology (2024)