Key Points
-
The intestinal microbiota coexists with its host in a continuum between homeostasis and pathogenicity; the upper gastrointestinal tract harbours a gut microbiota that is affected compositionally and metabolically by food components
-
Coeliac disease is a chronic immune-mediated enteropathy caused by dietary gluten in genetically susceptible individuals
-
The role of microbial factors in coeliac disease pathogenesis has been suggested
-
Although clinical studies demonstrate that microbial changes are associated with coeliac disease, the individual microbes involved and underlying mechanisms remain elusive
-
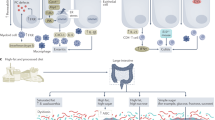
Emerging data in gnotobiotic models indicate that the intestinal microbiota has a complex modulatory role in host immune responses to gluten
-
A deeper understanding of the precise role of microbes in coeliac disease pathogenesis will aid in the development of microbiota-modulating strategies, such as probiotics, to prevent or help treat the disease
Abstract
Several studies point towards alteration in gut microbiota composition and function in coeliac disease, some of which can precede the onset of disease and/or persist when patients are on a gluten-free diet. Evidence also exists that the gut microbiota might promote or reduce coeliac-disease-associated immunopathology. However, additional studies are required in humans and in mice (using gnotobiotic technology) to determine cause–effect relationships and to identify agents for modulating the gut microbiota as a therapeutic or preventative approach for coeliac disease. In this Review, we summarize the current evidence for altered gut microbiota composition in coeliac disease and discuss how the interplay between host genetics, environmental factors and the intestinal microbiota might contribute to its pathogenesis. Moreover, we highlight the importance of utilizing animal models and long-term clinical studies to gain insight into the mechanisms through which host–microbial interactions can influence host responses to gluten.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Pabst, O. & Mowat, A. Oral tolerance to food protein. Mucosal Immunol. 5, 232–239 (2012).
Sommer, F. & Bäckhed, F. The gut microbiota—masters of host development and physiology. Nat. Rev. Microbiol. 11, 227–238 (2013).
Belkaid, Y. & Hand, T. W. Role of the microbiota in immunity and inflammation. Cell 157, 121–141 (2014).
Catassi, C. et al. Natural history of celiac disease autoimmunity in a USA cohort followed since 1974. Ann. Med. 42, 530–538 (2010).
Ludvigsson, J. F. et al. Increasing incidence of celiac disease in a North American population. Am. J. Gastroenterol. 108, 818–824 (2013).
Murray, J. A. et al. Trends in the identification and clinical features of celiac disease in a North American community, 1950–2001 Clin. Gastroenterol. Hepatol. 1, 19–27 (2003).
White, L. E. et al. The rising incidence of celiac disease in Scotland. Pediatrics 132, e924–e931 (2013).
Trynka, G. et al. Dense genotyping identifies and localizes multiple common and rare variant association signals in celiac disease. Nat. Gen. 43, 1193–1201 (2011).
Parmar, A. et al. Association study of FUT2 (rs601338) with celiac disease and inflammatory bowel disease in the Finnish population. Tissue Antigens 80, 488–493 (2012).
Tong, M. et al. Reprograming of gut microbiome energy metabolism by the FUT2 Crohn's disease risk polymorphism. ISME J. 8, 2193–2206 (2014).
Mowat, A. M. & Agace, W. W. Regional specialization within the intestinal immune system. Nat. Rev. Immunol. 14, 667–685 (2014).
Zoetendal, E. G. et al. The human small intestinal microbiota is driven by rapid uptake and conversion of simple carbohydrates. ISME J. 6, 1415–1426 (2012).
El Aidy, S. et al. Temporal and spatial interplay of microbiota and intestinal mucosa drive establishment of immune homeostasis in conventionalized mice. Mucosal Immunol. 5, 567–579 (2012).
Natividad, J. M. et al. Differential induction of antimicrobial REGIII by the intestinal microbiota and Bifidobacterium breve NCC2950. Appl. Environ. Microbiol. 79, 7745–7754 (2013).
Ivanov, I. I. et al. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe 4, 337–349 (2008).
Geuking, M. B. et al. Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity 34, 794–806 (2011).
Helgeland, L., Vaage, J., Rolstad, B., Midtvedt, T. & Brandtzaeg, P. Microbial colonization influences composition and T-cell receptor Vβ repertoire of intraepithelial lymphocytes in rat intestine. Immunology 89, 494–501 (1996).
Imaoka, A., Matsumoto, S., Setoyama, H., Okada, Y. & Umesaki, Y. Proliferative recruitment of intestinal intraepithelial lymphocytes after microbial colonization of germ-free mice. Eur. J. Immunol. 26, 945–948 (1996).
Uematsu, S. et al. Regulation of humoral and cellular gut immunity by lamina propria dendritic cells expressing Toll-like receptor 5. Nat. Immunol. 9, 769–776 (2008).
Hapfelmeier, S. et al. Reversible microbial colonization of germ-free mice reveals the dynamics of IgA immune responses. Science 328, 1705–1709 (2010).
Ismail, A. S., Behrendt, C. L. & Hooper, L. V. Reciprocal interactions between commensal bacteria and γδ intraepithelial lymphocytes during mucosal injury. J. Immunol. 182, 3047–3054 (2009).
Ismail, A. S. et al. γδ intraepithelial lymphocytes are essential mediators of host–microbial homeostasis at the intestinal mucosal surface. Proc. Natl Acad. Sci. USA 108, 8743–8748 (2011).
Vaishnava, S. et al. The antibacterial lectin RegIIIγ promotes the spatial segregation of microbiota and host in the intestine. Science 334, 255–258 (2011).
Sanos, S. L. et al. RORγt and commensal microflora are required for the differentiation of mucosal interleukin 22–producing NKp46+ cells. Nat. Immunol. 10, 83–91 (2009).
Chen, V. L. & Kasper, D. L. Interactions between the intestinal microbiota and innate lymphoid cells. Gut Microbes 5, 129–140 (2014).
Moro, K. & Koyasu, S. Innate lymphoid cells, possible interaction with microbiota. Semin. Immunopathol. 37, 27–37 (2015).
Kinnebrew, M. A. et al. Interleukin 23 production by intestinal CD103+ CD11b+ dendritic cells in response to bacterial flagellin enhances mucosal innate immune defense. Immunity 36, 276–287 (2012).
Ramos, H. C., Rumbo, M. & Sirard, J.-C. Bacterial flagellins: mediators of pathogenicity and host immune responses in mucosa. Trends Microbiol. 12, 509–517 (2004).
Duan, Q., Zhou, M., Zhu, L. & Zhu, G. Flagella and bacterial pathogenicity. J. Basic Microbiol. 53, 1–8 (2013).
Gaboriau-Routhiau, V. et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity 31, 677–689 (2009).
Ivanov, I. I. et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139, 485–498 (2009).
Atarashi, K. et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 500, 232–236 (2013).
Atarashi, K. et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science 331, 337–341 (2011).
Round, J. L. & Mazmanian, S. K. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc. Natl Acad. Sci. USA 107, 12204–12209 (2010).
Hrncir, T., Stepankova, R., Kozakova, H., Hudcovic, T. & Tlaskalova-Hogenova, H. Gut microbiota and lipopolysaccharide content of the diet influence development of regulatory T cells: studies in germ-free mice. BMC Immunol. 9, 65 (2008).
Arpaia, N. et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 504, 451–455 (2013).
Furusawa, Y. et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 504, 446–450 (2013).
Smith, P. M. et al. The microbial metabolites, short-chain fatty acids, regulate colonic TREG cell homeostasis. Science 341, 569–573 (2013).
Singh, N. et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity 40, 128–139 (2014).
McLean, M. H., Dieguez, D., Miller, L. M. & Young, H. A. Does the microbiota play a role in the pathogenesis of autoimmune diseases? Gut 64, 332–341 (2014).
Rossi, M. & Schwartz, K. B. Editorial: Celiac disease and intestinal bacteria: not only gluten? J. Leukoc. Biol. 87, 749–751 (2010).
Sánchez, E., Donat, E., Ribes-Koninckx, C., Fernández-Murga, M. L. & Sanz, Y. Duodenal-mucosal bacteria associated with celiac disease in children. Appl. Environ. Microbiol. 79, 5472–5479 (2013).
Wacklin, P. et al. The duodenal microbiota composition of adult celiac disease patients is associated with the clinical manifestation of the disease. Inflamm. Bowel Dis. 19, 934–941 (2013).
Collado, M. C., Donat, E., Ribes-Koninckx, C., Calabuig, M. & Sanz, Y. Specific duodenal and faecal bacterial groups associated with paediatric coeliac disease. J. Clin. Pathol. 62, 264–269 (2009).
Collado, M., Donat, E., Ribes-Koninckx, C., Calabuig, M. & Sanz, Y. Imbalances in faecal and duodenal Bifidobacterium species composition in active and non-active coeliac disease. BMC Microbiol. 8, 232 (2008).
Di Cagno, R. et al. Duodenal and faecal microbiota of celiac children: molecular, phenotype and metabolome characterization. BMC Microbiol. 11, 219 (2011).
De Palma, G. et al. Intestinal dysbiosis and reduced immunoglobulin-coated bacteria associated with coeliac disease in children. BMC Microbiol. 10, 63 (2010).
Sanz, Y. et al. Differences in faecal bacterial communities in coeliac and healthy children as detected by PCR and denaturing gradient gel electrophoresis. FEMS Immunol. Med. Microbiol. 51, 562–568 (2007).
Nadal, I., Donant, E., Ribes-Koninckx, C., Calabuig, M. & Sanz, Y. Imbalance in the composition of the duodenal microbiota of children with coeliac disease. J. Med. Microbiol. 56, 1669–1674 (2007).
Schippa, S. et al. A distinctive 'microbial signature'in celiac pediatric patients. BMC Microbiol. 10, 175 (2010).
Wacklin, P. et al. Altered duodenal microbiota composition in celiac disease patients suffering from persistent symptoms on a long-term gluten-free diet. Am. J. Gastroenterol. 109, 1933–1941 (2014).
Ivarsson, A. et al. Epidemic of coeliac disease in Swedish children. Acta Paediatr. 89, 165–171 (2000).
Ou, G. et al. Proximal small intestinal microbiota and identification of rod-shaped bacteria associated with childhood celiac disease. Am. J. Gastroenterol. 104, 3058–3067 (2009).
Decker, E. et al. Cesarean delivery is associated with celiac disease but not inflammatory bowel disease in children. Pediatrics 125, e1433–e1440 (2010).
Mårild, K., Stephansson, O., Montgomery, S., Murray, J. A. & Ludvigsson, J. F. Pregnancy outcome and risk of celiac disease in offspring: a nationwide case-control study. Gastroenterology 142, 39–45 (2012).
Emilsson, L., Magnus, M. C. & Størdal, K. Perinatal risk factors for development of celiac disease in children, based on the prospective Norwegian Mother and Child cohort study. Clin. Gastroenterol. Hepatol. 13, 921–927 (2015).
Canova, C. et al. Association of maternal education, early infections, and antibiotic use with celiac disease: a population-based birth cohort study in northeastern Italy. Am. J. Epidemiol. 180, 76–85 (2014).
Ivarsson, A. et al. Prevalence of childhood celiac disease and changes in infant feeding. Pediatrics 131, e687–e694 (2013).
Lionetti, E. et al. Introduction of gluten, HLA status, and the risk of celiac disease in children. N. Engl. J. Med. 371, 1295–1303 (2014).
Vriezinga, S. L. et al. Randomized feeding intervention in infants at high risk for celiac disease. N. Engl. J. Med. 371, 1304–1315 (2014).
Jansen, M. A. et al. Infant feeding and anti-tissue transglutaminase antibody concentrations in the Generation R Study. Am. J. Clin. Nutr. 100, 1095–1101 (2014).
Collins, S. M. A role for the gut microbiota in IBS. Nat. Rev. Gastroenterol. Hepatol. 11, 497–505 (2014).
Matsuoka, K. & Kanai, T. The gut microbiota and inflammatory bowel disease. Semin. Immunopathol. 37, 47–55 (2015).
Manichanh, C., Borruel, N., Casellas, F. & Guarner, F. The gut microbiota in IBD. Nat. Rev. Gastroenterol. Hepatol. 9, 599–608 (2012).
Rajilic´-Stojanovic´, M. et al. Intestinal microbiota and diet in IBS: causes, consequences, or epiphenomena? Am. J. Gastroenterol. 110, 278–287 (2015).
Simrén, M. et al. Intestinal microbiota in functional bowel disorders: a Rome foundation report. Gut 62, 159–176 (2012).
Walters, W. A., Xu, Z. & Knight, R. Meta-analyses of human gut microbes associated with obesity and IBD. FEBS Lett. 588, 4223–4233 (2014).
Tims, S. et al. Microbiota conservation and BMI signatures in adult monozygotic twins. ISME J. 7, 707–717 (2013).
Turnbaugh, P. J. et al. A core gut microbiome in obese and lean twins. Nature 457, 480–484 (2009).
Yatsunenko, T. et al. Human gut microbiome viewed across age and geography. Nature 486, 222–227 (2012).
Frank, D. N. et al. Disease phenotype and genotype are associated with shifts in intestinal-associated microbiota in inflammatory bowel diseases. Inflamm. Bowel Dis. 17, 179–184 (2011).
Khachatryan, Z. A. et al. Predominant role of host genetics in controlling the composition of gut microbiota. PLoS ONE 3, e3064 (2008).
Rausch, P. et al. Colonic mucosa-associated microbiota is influenced by an interaction of Crohn disease and FUT2 (Secretor) genotype. Proc. Natl Acad. Sci. USA 108, 19030–19035 (2011).
Rehman, A. et al. Nod2 is essential for temporal development of intestinal microbial communities. Gut 60, 1354–1362 (2011).
Goodrich, J. K. et al. Human genetics shape the gut microbiome. Cell 159, 789–799 (2014).
Benson, A. K. et al. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc. Natl Acad. Sci. USA 107, 18933–18938 (2010).
McKnite, A. M. et al. Murine gut microbiota is defined by host genetics and modulates variation of metabolic traits. PLoS ONE 7, e39191 (2012).
Spor, A., Koren, O. & Ley, R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat. Rev. Microbiol. 9, 279–290 (2011).
Natividad, J. M. M. et al. Commensal and probiotic bacteria influence intestinal barrier function and susceptibility to colitis in Nod1−/−; Nod2−/− mice. Inflamm. Bowel Dis. 18, 1434–1446 (2012).
Robertson, S. J. et al. Nod1 and Nod2 signaling does not alter the composition of intestinal bacterial communities at homeostasis. Gut Microbes 4, 222–231 (2013).
Ubeda, C. et al. Familial transmission rather than defective innate immunity shapes the distinct intestinal microbiota of TLR-deficient mice. J. Exp. Med. 209, 1445–1456 (2012).
Carmody, R. N. et al. Diet dominates host genotype in shaping the murine gut microbiota. Cell Host Microbe 17, 72–84 (2015).
Toivanen, P., Vaahtovuo, J. & Eerola, E. Influence of major histocompatibility complex on bacterial composition of fecal flora. Infect. Immun. 69, 2372–2377 (2001).
Palma, G. d., Nova, E., Pozo, T. & Sanz, Y. Interplay between human leukocyte antigen genes and the microbial colonization process of the newborn intestine. Curr. Issues Mol. Biol. 12, 1–10 (2010).
Sánchez, E. et al. Influence of environmental and genetic factors linked to celiac disease risk on infant gut colonization by Bacteroides species. Appl. Environ. Microbiol. 77, 5316–5323 (2011).
De Palma, G. et al. Influence of milk-feeding type and genetic risk of developing coeliac disease on intestinal microbiota of infants: the PROFICEL study. PLoS ONE 7, e30791 (2012).
Sellitto, M. et al. Proof of concept of microbiome-metabolome analysis and delayed gluten exposure on celiac disease autoimmunity in genetically at-risk infants. PLoS ONE 7, e33387 (2012).
Olivares, M. et al. The HLA-DQ2 genotype selects for early intestinal microbiota composition in infants at high risk of developing coeliac disease. Gut 64, 406–417 (2014).
Wacklin, P. et al. Secretor genotype (FUT2 gene) is strongly associated with the composition of Bifidobacteria in the human intestine. PLoS ONE 6, e20113 (2011).
Stapleton, A., Nudelman, E., Clausen, H., Hakomori, S.-I. & Stamm, W. Binding of uropathogenic Escherichia coli R45 to glycolipids extracted from vaginal epithelial cells is dependent on histo-blood group secretor status. J. Clin. Invest. 90, 965–972 (1992).
De Palma, G., Cinova, J., Stepankova, R., Tuckova, L. & Sanz, Y. Pivotal Advance: Bifidobacteria and Gram-negative bacteria differentially influence immune responses in the proinflammatory milieu of celiac disease. J. Leukoc. Biol. 87, 765–778 (2010).
De Palma, G. et al. Modulation of phenotypic and functional maturation of dendritic cells by intestinal bacteria and gliadin: relevance for celiac disease. J. Leukoc. Biol. 92, 1043–1054 (2012).
Laparra, J. M. & Sanz, Y. Bifidobacteria inhibit the inflammatory response induced by gliadins in intestinal epithelial cells via modifications of toxic peptide generation during digestion. J. Cell. Biochem. 109, 801–807 (2010).
Lindfors, K. et al. Live probiotic Bifidobacterium lactis bacteria inhibit the toxic effects induced by wheat gliadin in epithelial cell culture. Clin. Exp. Immunol. 152, 552–558 (2008).
Cinova, J. et al. Role of intestinal bacteria in gliadin-induced changes in intestinal mucosa: study in germ-free rats. PLoS ONE 6, e16169 (2011).
Štepánková, R., Tlaskalova-Hogenova, H., Šinkora, J., Jodl, J. & Fric, P. Changes in jejunal mucosa after long-term feeding of germfree rats with gluten. Scand. J. Gastroenterol. 31, 551–557 (1996).
Galipeau, H. J. et al. Sensitization to gliadin induces moderate enteropathy and insulitis in nonobese diabetic-DQ8 mice. J. Immunol. 187, 4338–4346 (2011).
Galipeau, H. et al. Gluten-induced responses in NOD/DQ8 mice are influenced by bacterial colonization [abstract Tu1749]. Gastroenterology 146 (Suppl. 1), S833 (2014).
Roberts, A. I. et al. Cutting edge: NKG2D receptors induced by IL-15 costimulate CD28-negative effector CTL in the tissue microenvironment. J. Immunol. 167, 5527–5530 (2001).
Meresse, B. et al. Coordinated induction by IL15 of a TCR-independent NKG2D signaling pathway converts CTL into lymphokine-activated killer cells in celiac disease. Immunity 21, 357–366 (2004).
Tang, F. et al. Cytosolic PLA2 is required for CTL-mediated immunopathology of celiac disease via NKG2D and IL-15. J. Exp. Med. 206, 707–719 (2009).
Mention, J. J. et al. Interleukin 15: a key to disrupted intraepithelial lymphocyte homeostasis and lymphomagenesis in celiac disease. Gastroenterology 125, 730–745 (2003).
Malamut, G. et al. IL-15 triggers an antiapoptotic pathway in human intraepithelial lymphocytes that is a potential new target in celiac disease–associated inflammation and lymphomagenesis. J. Clin. Invest. 120, 2131–2143 (2010).
DePaolo, R. W. et al. Co-adjuvant effects of retinoic acid and IL-15 induce inflammatory immunity to dietary antigens. Nature 471, 220–224 (2011).
Benahmed, M. et al. Inhibition of TGF-β signaling by IL-15: a new role for IL-15 in the loss of immune homeostasis in celiac disease. Gastroenterology 132, 994–1008 (2007).
Hmida, N. B. et al. Impaired control of effector T cells by regulatory T cells: a clue to loss of oral tolerance and autoimmunity in celiac disease. Am. J. Gastroenterol. 107, 604–611 (2011).
Zhang, X., Sun, S., Hwang, I., Tough, D. F. & Sprent, J. Potent and selective stimulation of memory-phenotype CD8+ T cells in vivo by IL-15. Immunity 8, 591–599 (1998).
Mattei, F., Schiavoni, G., Belardelli, F. & Tough, D. F. IL-15 is expressed by dendritic cells in response to type I IFN, double-stranded RNA, or lipopolysaccharide and promotes dendritic cell activation. J. Immunol. 167, 1179–1187 (2001).
Korneychuk, N., Meresse, B. & Cerf-Bensussan, N. Lessons from rodent models in celiac disease. Mucosal Immunol. 8, 18–28 (2015).
Olivares, M., Castillejo, G., Varea, V. & Sanz, Y. Double-blind, randomised, placebo-controlled intervention trial to evaluate the effects of Bifidobacterium longum CECT 7347 in children with newly diagnosed coeliac disease. Br. J. Nutr. 112, 30–40 (2014).
Smecuol, E. et al. Exploratory, randomized, double-blind, placebo-controlled study on the effects of Bifidobacterium infantis natren life start strain super strain in active celiac disease. J. Clin. Gastroenterol. 47, 139–147 (2013).
Batt, R., Carter, M. & McLean, L. Morphological and biochemical studies of a naturally occurring enteropathy in the Irish setter dog: a comparison with coeliac disease in man. Res. Vet. Sci. 37, 339–346 (1984).
Hall, E. & Batt, R. Dietary modulation of gluten sensitivity in a naturally occurring enteropathy of Irish setter dogs. Gut 33, 198–205 (1992).
Polvi, A. et al. Genetic susceptibility to gluten sensitive enteropathy in Irish setter dogs is not linked to the major histocompatibility complex. Tissue Antigens 52, 543–549 (1998).
Sestak, K. et al. Recognition of epidermal transglutaminase by IgA and tissue transglutaminase 2 antibodies in a rare case of rhesus dermatitis. J. Vis. Exp. 58, e3154 (2011).
Bethune, M. T. et al. A non-human primate model for gluten sensitivity. PLoS ONE 3, e1614 (2008).
van der Kolk, J. et al. Gluten-dependent antibodies in horses with inflammatory small bowel disease (ISBD). Vet. Q. 32, 3–11 (2012).
Papista, C. et al. Gluten induces coeliac-like disease in sensitised mice involving IgA, CD71 and transglutaminase 2 interactions that are prevented by probiotics. Lab. Invest. 92, 625–635 (2012).
Freitag, T. et al. Gliadin-primed CD4+CD45RBlowCD25-effector/memory T cells drive gluten-dependent small intestinal damage after adoptive transfer into lymphopenic mice. Gut 58, 1597–1605 (2009).
Black, K. E., Murray, J. A. & David, C. S. HLA-DQ determines the response to exogenous wheat proteins: a model of gluten sensitivity in transgenic knockout mice. J. Immunol. 169, 5595–5600 (2002).
Verdu, E. F. et al. Gliadin-dependent neuromuscular and epithelial secretory responses in gluten-sensitive HLA-DQ8 transgenic mice. Am J. Physiol. Gastrointest. Liver Physiol. 294, G217–G225 (2008).
de Kauwe, A. L. et al. Resistance to celiac disease in humanized HLA-DR3-DQ2-transgenic mice expressing specific anti-gliadin CD4+ T cells. J. Immunol. 182, 7440–7450 (2009).
Maurano, F. et al. Small intestinal enteropathy in non-obese diabetic mice fed a diet containing wheat. Diabetologia 48, 931–937 (2005).
Acknowledgements
The authors are supported by grants from the Canadian Institutes of Health Research to E.F.V. (MOP 123282); and the Digestive Diseases Research Core Centre (DK42086) at the University of Chicago and from the NIH (RO1DK67180) to B.J. E.F.V. holds a Canada Research Chair and H.J.G. a Canadian Celiac Association fellowship.
Author information
Authors and Affiliations
Contributions
All authors contributed equally to all aspects of this manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Table 1
Summary of microbial alterations in patients with coeliac disease (DOC 117 kb)
Rights and permissions
About this article
Cite this article
Verdu, E., Galipeau, H. & Jabri, B. Novel players in coeliac disease pathogenesis: role of the gut microbiota. Nat Rev Gastroenterol Hepatol 12, 497–506 (2015). https://doi.org/10.1038/nrgastro.2015.90
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrgastro.2015.90
This article is cited by
-
Polymeric immunoglobulin receptor deficiency exacerbates autoimmune hepatitis by inducing intestinal dysbiosis and barrier dysfunction
Cell Death & Disease (2023)
-
The impact of mass drug administration of antibiotics on the gut microbiota of target populations
Infectious Diseases of Poverty (2022)
-
Maternal breast milk microbiota and immune markers in relation to subsequent development of celiac disease in offspring
Scientific Reports (2022)
-
Spectroscopic investigation of faeces with surface-enhanced Raman scattering: a case study with coeliac patients on gluten-free diet
Analytical and Bioanalytical Chemistry (2022)
-
Role of gene regulation and inter species interaction as a key factor in gut microbiota adaptation
Archives of Microbiology (2022)