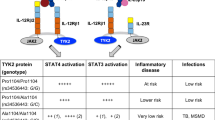
Key Points
-
The Immunochip project has identified >160 loci containing IBD genes and 70% of these loci are shared with other immune-mediated inflammatory diseases
-
Similar to other complex polygenic diseases, only a small fraction of heritability is explained by the genetic loci identified in IBD
-
Hypothesis-free genetic association studies in IBD have identified key pathways involved in innate immunity, autophagy, lymphocyte differentiation and chemotaxis
-
Genetic corroboration is now available for novel treatment strategies targeting IL-12–IL-23 signalling, JAK–STAT signalling and leukocyte chemotaxis
-
The paradigm of personalized IBD care will probably only be achieved if we succeed in integrating the genetic and basic science advances with insights into the ecology of the gut microbiota
Abstract
IBD is a spectrum of chronic disorders that constitute an important health problem worldwide. The hunt for genetic determinants of disease onset and course has culminated in the Immunochip project, which has identified >160 loci containing IBD susceptibility genes. In this Review, we highlight how genetic association studies have informed our understanding of the pathogenesis of IBD by focusing research efforts on key pathways involved in innate immunity, autophagy, lymphocyte differentiation and chemotaxis. Several of these novel genetic markers and cellular pathways are promising candidates for patient stratification and therapeutic targeting.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Molodecky, N. A. et al. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology 142, 46–54 (2012).
Jess, T., Frisch, M. & Simonsen, J. Trends in overall and cause-specific mortality among patients with inflammatory bowel disease from 1982 to 2010. Clin. Gastroenterol. Hepatol. 11, 43–48 (2013).
van Heel, D. A. et al. Inflammatory bowel disease susceptibility loci defined by genome scan meta-analysis of 1952 affected relative pairs. Hum. Mol. Genet. 13, 763–770 (2004).
Van Limbergen, J. et al. Definition of phenotypic characteristics of childhood-onset inflammatory bowel disease. Gastroenterology 135, 1114–1122 (2008).
Satsangi, J., Kennedy, N. A., Henderson, P., Wilson, D. C. & Nimmo, E. R. Exploring the hidden heritability of inflammatory bowel disease. Gut 60, 1447–1448 (2011).
Eichler, E. E. et al. Missing heritability and strategies for finding the underlying causes of complex disease. Nat. Rev. Genet. 11, 446–450 (2010).
Halfvarson, J. Genetics in twins with Crohn's disease: less pronounced than previously believed? Inflamm. Bowel Dis. 17, 6–12 (2011).
Zuk, O., Hechter, E., Sunyaev, S. R. & Lander, E. S. The mystery of missing heritability: genetic interactions create phantom heritability. Proc. Natl Acad. Sci. USA 109, 1193–1198 (2012).
Dunham, I. et al. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012).
Parkes, M., Cortes, A., van Heel, D. A. & Brown, M. A. Genetic insights into common pathways and complex relationships among immune-mediated diseases. Nat. Rev. Genet. 14, 661–673 (2013).
Zheng, Y. et al. Role of conserved non-coding DNA elements in the Foxp3 gene in regulatory T-cell fate. Nature 463, 808–812 (2010).
Muise, A. M., Snapper, S. B. & Kugathasan, S. The age of gene discovery in very early onset inflammatory bowel disease. Gastroenterology 143, 285–288 (2012).
Christodoulou, K. et al. Next generation exome sequencing of paediatric inflammatory bowel disease patients identifies rare and novel variants in candidate genes. Gut 62, 977–984 (2013).
Muise, A. M. et al. NADPH oxidase complex and IBD candidate gene studies: identification of a rare variant in NCF2 that results in reduced binding to RAC2. Gut 61, 1028–1035 (2012).
Tyler, A. D. et al. The NOD2insC polymorphism is associated with worse outcome following ileal pouch-anal anastomosis for ulcerative colitis. Gut 62, 1433–1439 (2013).
Eglinton, T. W. et al. Clinical and genetic risk factors for perianal Crohn's disease in a population-based cohort. Am. J. Gastroenterol. 107, 589–596 (2012).
Speckmann, C. & Ehl, S. XIAP deficiency is a mendelian cause of late-onset IBD. Gut http://dx.doi.org/10.1136/gutjnl-2013-306474.
Avitzur, Y. et al. Mutations in tetratricopeptide repeat domain 7A result in a severe form of very early onset inflammatory bowel disease. Gastroenterology http://dx.doi.org/10.1053/j.gastro.2014.01.015.
Uhlig, H. H. Monogenic diseases associated with intestinal inflammation: implications for the understanding of inflammatory bowel disease. Gut 62, 1795–1805 (2013).
NEOPICS. NEOPICS [online], (2014).
Momozawa, Y. et al. Resequencing of positional candidates identifies low frequency IL23R coding variants protecting against inflammatory bowel disease. Nat. Genet. 43, 43–47 (2011).
Rivas, M. A. et al. Deep resequencing of GWAS loci identifies independent rare variants associated with inflammatory bowel disease. Nat. Genet. 43, 1066–1073 (2011).
Jostins, L. et al. Host–microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 491, 119–124 (2012).
Hunt, K. A. et al. Negligible impact of rare autoimmune-locus coding-region variants on missing heritability. Nature 498, 232–235 (2013).
Beaudoin, M. et al. Deep resequencing of GWAS loci identifies rare variants in CARD9, IL23R and RNF186 that are associated with ulcerative colitis. PLoS Genet. 9, e1003723 (2013).
Ellinghaus, D. et al. Association between variants of PRDM1 and NDP52 and Crohn's disease, based on exome sequencing and functional studies. Gastroenterology 145, 339–347 (2013).
Wang, K. et al. Diverse genome-wide association studies associate the IL12/IL23 pathway with Crohn Disease. Am. J. Hum. Genet. 84, 399–405 (2009).
Van Limbergen, J., Philpott, D. & Griffiths, A. M. Genetic profiling in inflammatory bowel disease: from association to bedside. Gastroenterology 141, 1566–1571 (2011).
Rossin, E. J. et al. Proteins encoded in genomic regions associated with immune-mediated disease physically interact and suggest underlying biology. PLoS Genet. 7, e1001273 (2011).
Ng, S. C. et al. Geographical variability and environmental risk factors in inflammatory bowel disease. Gut 62, 630–649 (2013).
Wang, M. H. et al. Gene–gene and gene–environment interactions in ulcerative colitis. Hum. Genet. http://dx.doi.org/10.1007/s00439-013-1395-z.
Hansen, R. et al. Microbiota of de-novo pediatric IBD: increased Faecalibacterium prausnitzii and reduced bacterial diversity in Crohn's but not in ulcerative colitis. Am. J. Gastroenterol. 107, 1913–1922 (2012).
Small, C. L., Reid-Yu, S. A., McPhee, J. B. & Coombes, B. K. Persistent infection with Crohn's disease-associated adherent-invasive Escherichia coli leads to chronic inflammation and intestinal fibrosis. Nat. Commun. 4, 1957 (2013).
Machiels, K. et al. A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut http://dx.doi.org/10.1136/gutjnl-2013-304833.
Boon, E. et al. Interactions in the microbiome: communities of organisms and communities of genes. FEMS Microbiol. Rev. 38, 90–118 (2013).
Chassaing, B., Koren, O., Carvalho, F. A., Ley, R. E. & Gewirtz, A. T. AIEC pathobiont instigates chronic colitis in susceptible hosts by altering microbiota composition. Gut http://dx.doi.org/10.1136/gutjnl-2013-304909.
Hooper, L. V., Littman, D. R. & Macpherson, A. J. Interactions between the microbiota and the immune system. Science 336, 1268–1273 (2012).
Biedermann, L. et al. Smoking cessation induces profound changes in the composition of the intestinal microbiota in humans. PLoS ONE 8, e59260 (2013).
Li, E. et al. Inflammatory bowel diseases phenotype, C. difficile and NOD2 genotype are associated with shifts in human ileum associated microbial composition. PLoS ONE 7, e26284 (2012).
Wang, M.-H. et al. A novel approach to detect cumulative genetic effects and genetic interactions in Crohn's Disease. Inflamm. Bowel Dis. 19, 1799–1808 (2013).
Cleynen, I. et al. Genetic factors conferring an increased susceptibility to develop Crohn's disease also influence disease phenotype: results from the IBDchip European Project. Gut 62, 1556–1565 (2012).
Barrett, J. C. et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat. Genet. 40, 955–962 (2008).
Van Limbergen, J., Wilson, D. C. & Satsangi, J. The genetics of Crohn's disease. Annu. Rev. Genomics Hum. Genet. 10, 89–116 (2009).
Imielinski, M. et al. Common variants at five new loci associated with early-onset inflammatory bowel disease. Nat. Genet. 41, 1335–1340 (2009).
Sherlock, M. E. et al. Infliximab-induced psoriasis and psoriasiform skin lesions in pediatric Crohn disease and a potential association with IL-23 receptor polymorphisms. J. Pediatr. Gastroenterol. Nutr. 56, 512–518 (2013).
Cleynen, I. & Vermeire, S. Paradoxical inflammation induced by anti-TNF agents in patients with IBD. Nat. Rev. Gastroenterol. Hepatol. 9, 496–503 (2012).
Clarke, L. et al. The 1000 Genomes Project: data management and community access. Nat. Methods 9, 459–462 (2012).
Hugot, J. P. et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature 411, 599–603 (2001).
Duerr, R. H. et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science 314, 1461–1463 (2006).
Hampe, J. et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat. Genet. 39, 207–211 (2007).
Van Limbergen, J. et al. Haplotype-tagging analysis of common variants of the IL23R gene demonstrates gene-wide extent of association with IBD. Inflamm. Bowel Dis. 19, E79–E80 (2013).
Van Limbergen, J. et al. Hypothesis-free analysis of ATG16L1 demonstrates gene-wide extent of association with Crohn's disease susceptibility. Gut 62, 331–333 (2013).
Raychaudhuri, S. et al. Identifying relationships among genomic disease regions: predicting genes at pathogenic SNP associations and rare deletions. PLoS Genet. 5, e1000534 (2009).
Lee, I., Blom, U. M., Wang, P. I., Shim, J. E. & Marcotte, E. M. Prioritizing candidate disease genes by network-based boosting of genome-wide association data. Genome Res. 21, 1109–1121 (2011).
Sorbara, M. T. et al. The protein ATG16L1 suppresses inflammatory cytokines induced by the intracellular sensors Nod1 and Nod2 in an autophagy-independent manner. Immunity 39, 858–873 (2013).
Ogura, Y. et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature 411, 603–606 (2001).
Girardin, S. E. et al. Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J. Biol. Chem. 278, 8869–8872 (2003).
Sabbah, A. et al. Activation of innate immune antiviral responses by Nod2. Nat. Immunol. 10, 1073–1080 (2009).
Adler, J., Rangwalla, S. C., Dwamena, B. A. & Higgins, P. D. The prognostic power of the NOD2 genotype for complicated Crohn's disease: a meta-analysis. Am. J. Gastroenterol. 106, 699–712 (2011).
Gutierrez, A. et al. Genetic susceptibility to increased bacterial translocation influences the response to biological therapy in patients with Crohn's disease. Gut 63, 272–280 (2014).
Mo, J. et al. Pathogen sensing by nucleotide-binding oligomerization domain-containing protein 2 (NOD2) is mediated by direct binding to muramyl dipeptide and ATP. J. Biol. Chem. 287, 23057–23067 (2012).
Coulombe, F. et al. Increased NOD2-mediated recognition of N-glycolyl muramyl dipeptide. J. Exp. Med. 206, 1709–1716 (2009).
Lesage, S. et al. CARD15/NOD2 mutational analysis and genotype–phenotype correlation in 612 patients with inflammatory bowel disease. Am. J. Hum. Genet. 70, 845–857 (2002).
Couturier-Maillard, A. et al. NOD2-mediated dysbiosis predisposes mice to transmissible colitis and colorectal cancer. J. Clin. Invest. 123, 700–711 (2013).
Schirbel, A. et al. Pro-angiogenic activity of TLRs and NLRs: a novel link between gut microbiota and intestinal angiogenesis. Gastroenterology 144, 613–623 (2013).
Kobayashi, K. S. et al. Nod2-dependent regulation of innate and adaptive immunity in the intestinal tract. Science 307, 731–734 (2005).
Bevins, C. L., Stange, E. F. & Wehkamp, J. Decreased Paneth cell defensin expression in ileal Crohn's disease is independent of inflammation, but linked to the NOD2 1007fs genotype. Gut 58, 882–883 (2009).
Simms, L. A. et al. Reduced α-defensin expression is associated with inflammation and not NOD2 mutation status in ileal Crohn's disease. Gut 57, 903–910 (2008).
Shanahan, M. T. et al. Mouse Paneth cell antimicrobial function is independent of Nod2. Gut http://dx.doi.org/10.1136/gutjnl-2012-304190.
Lipinski, S. et al. RNAi screening identifies mediators of NOD2 signaling: Implications for spatial specificity of MDP recognition. Proc. Natl Acad. Sci U.S.A. 109, 21426–21431 (2012).
Philpott, D. J., Sorbara, M. T., Roberston, S. J., Croitoru, K. & Girardin, S. E. NOD proteins: regulators of inflammation in health and disease. Nat. Rev. Immunol. 14, 9–23 (2014).
Billmann-Born, S. et al. Genome-wide expression profiling identifies an impairment of negative feedback signals in the Crohn's disease-associated NOD2 variant L1007fsinsC. J. Immunol. 186, 4027–4038 (2011).
Corridoni, D. et al. Dysregulated NOD2 predisposes SAMP1/YitFc mice to chronic intestinal inflammation. Proc. Natl Acad. Sci. USA 110, 16999–17004 (2013).
Noguchi, E., Homma, Y., Kang, X., Netea, M. G. & Ma, X. A Crohn's disease-associated NOD2 mutation suppresses transcription of human IL10 by inhibiting activity of the nuclear ribonucleoprotein hnRNP-A1. Nat. Immunol. 10, 471–479 (2009).
Nimmo, E. R. et al. TLE1 modifies the effects of NOD2 in the pathogenesis of Crohn's disease. Gastroenterology 141, 972–981. e1–e2 (2011).
Spalinger, M. R. et al. Protein tyrosine phosphatase non-receptor type 22 modulates NOD2-induced cytokine release and autophagy. PLoS ONE 8, e72384 (2013).
Ghorpade, D. S., Kaveri, S. V., Bayry, J. & Balaji, K. N. Cooperative regulation of NOTCH1 protein-phosphatidylinositol 3-kinase (PI3K) signaling by NOD1, NOD2, and TLR2 receptors renders enhanced refractoriness to transforming growth factor-β (TGF-β)- or cytotoxic T-lymphocyte antigen 4 (CTLA-4)-mediated impairment of human dendritic cell maturation. J. Biol. Chem. 286, 31347–31360 (2011).
Cheon, J. H. Genetics of inflammatory bowel diseases: A comparison between Western and Eastern perspectives. J. Gastroenterol. Hepatol. 28, 220–226 (2013).
Brain, O. et al. The intracellular sensor NOD2 induces microRNA-29 expression in human dendritic cells to limit IL-23 release. Immunity 39, 521–536 (2013).
Stevens, C. et al. The intermediate filament protein, vimentin, is a regulator of NOD2 activity. Gut 62, 695–707 (2013).
Krishnaswamy, J. K., Chu, T. & Eisenbarth, S. C. Beyond pattern recognition: NOD-like receptors in dendritic cells. Trends Immunol. 34, 224–233 (2013).
Travassos, L. H. et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat. Immunol. 11, 55–62 (2010).
Stuart, L. M., Paquette, N. & Boyer, L. Effector-triggered versus pattern-triggered immunity: how animals sense pathogens. Nat. Rev. Immunol. 13, 199–206 (2013).
Boyer, L. et al. Pathogen-derived effectors trigger protective immunity via activation of the Rac2 enzyme and the IMD or Rip kinase signaling pathway. Immunity 35, 536–549 (2011).
Zhernakova, A. et al. Genetic analysis of innate immunity in Crohn's disease and ulcerative colitis identifies two susceptibility loci harboring CARD9 and IL18RAP. Am. J. Hum. Genet. 82, 1202–1210 (2008).
Cooke, J. et al. Mucosal genome-wide methylation changes in inflammatory bowel disease. Inflamm. Bowel Dis. 18, 2128–2137 (2012).
Gross, O. Card9 controls a non-TLR signalling pathway for innate anti-fungal immunity. Nature 442, 651–656 (2006).
Dubois, P. C. et al. Multiple common variants for celiac disease influencing immune gene expression. Nat. Genet. 42, 295–302 (2010).
Trynka, G. et al. Dense genotyping identifies and localizes multiple common and rare variant association signals in celiac disease. Nat. Genet. 43, 1193–1201 (2011).
Tsoi, L. C. et al. Identification of 15 new psoriasis susceptibility loci highlights the role of innate immunity. Nat. Genet. 44, 1341–1348 (2012).
Liu, H. et al. Identification of IL18RAP/IL18R1 and IL12B as leprosy risk genes demonstrates shared pathogenesis between inflammation and infectious diseases. Am. J. Hum. Genet. 91, 935–941 (2012).
Henckaerts, L. & Vermeire, S. NOD2/CARD15 disease associations other than Crohn's disease. Inflamm. Bowel Dis. 13, 235–241 (2007).
Kotlarz, D. et al. Loss of interleukin-10 signaling and infantile inflammatory bowel disease: implications for diagnosis and therapy. Gastroenterology 143, 347–355 (2012).
Stepensky, P. et al. Deficiency of caspase recruitment domain family, member 11 (CARD11), causes profound combined immunodeficiency in human subjects. J. Allergy Clin. Immunol. 131, 477–485. e1 (2013).
Molinero, L. L., Cubre, A., Mora-Solano, C., Wang, Y. & Alegre, M. L. T cell receptor/CARMA1/NF-κB signaling controls T-helper (Th) 17 differentiation. Proc. Natl Acad. Sci. USA 109, 18529–18534 (2012).
Glocker, E. O. et al. A homozygous CARD9 mutation in a family with susceptibility to fungal infections. N. Engl. J. Med. 361, 1727–1735 (2009).
Drewniak, A. A. et al. Invasive fungal infection and impaired neutrophil killing in human CARD9 deficiency. Blood 121, 2385–2392 (2013).
Hara, H. & Saito, T. CARD9 versus CARMA1 in innate and adaptive immunity. Trends Immunol. 30, 234–242 (2009).
Pelzer, C. et al. The protease activity of the paracaspase MALT1 is controlled by monoubiquitination. Nat. Immunol. 14, 337–345 (2013).
Pariente, B. et al. Activation of the receptor NKG2D leads to production of TH17 cytokines in CD4+ T cells of patients with Crohn's disease. Gastroenterology 141, 217–226 (2011).
Iliev, I. D. et al. Interactions between commensal fungi and the c-type lectin receptor dectin-1 influence colitis. Science 336, 1314–1317 (2012).
Arijs, I. et al. Mucosal gene expression of cell adhesion molecules, chemokines, and chemokine receptors in patients with inflammatory bowel disease before and after infliximab treatment. Am. J. Gastroenterol. 106, 748–761 (2011).
Wu, W., Hsu, Y. M., Bi, L., Songyang, Z. & Lin, X. CARD9 facilitates microbe-elicited production of reactive oxygen species by regulating the LyGDI–Rac1 complex. Nat. Immunol. 10, 1208–1214 (2009).
Gianni, T., Leoni, V., Chesnokova, L. S., Hutt-Fletcher, L. M. & Campadelli-Fiume, G. αvβ3-integrin is a major sensor and activator of innate immunity to herpes simplex virus-1. Proc. Natl Acad. Sci. USA 109, 19792–19797 (2012).
Hsu, Y. M. The adaptor protein CARD9 is required for innate immune responses to intracellular pathogens. Nat. Immunol. 8, 198–205 (2007).
Molinero, L. L. et al. CARMA1 controls an early checkpoint in the thymic development of FoxP3+ regulatory T cells. J. Immunol. 182, 6736–6743 (2009).
Rioux, J. D. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat. Genet. 39, 596–604 (2007).
Van Limbergen, J. E., Stevens, C., Nimmo, E. R., Wilson, D. C. & Satsangi, J. Autophagy: from basic science to clinical application. Mucosal Immunol. 2, 315–330 (2009).
Tattoli, I. et al. Amino acid starvation induced by invasive bacterial pathogens triggers an innate host defense program. Cell Host Microbe 11, 563–575 (2012).
Singh, S. B., Davis, A. S., Taylor, G. A. & Deretic, V. Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science 313, 1438–1441 (2006).
Orvedahl, A. et al. Image-based genome-wide siRNA screen identifies selective autophagy factors. Nature 480, 113–117 (2011).
Cadwell, K. et al. Virus-plus-susceptibility gene interaction determines Crohn's disease gene Atg16l1 phenotypes in intestine. Cell 141, 1135–1145 (2010).
Cadwell, K. et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature 456, 259–263 (2008).
Saitoh, T. et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1β production. Nature 456, 264–268 (2008).
Kuballa, P., Huett, A., Rioux, J. D., Daly, M. J. & Xavier, R. J. Impaired autophagy of an intracellular pathogen induced by a Crohn's disease associated ATG16L1 variant. PLoS ONE 3, e3391 (2008).
Plantinga, T. S. et al. Crohn's disease-associated ATG16L1 polymorphism modulates pro-inflammatory cytokine responses selectively upon activation of NOD2. Gut 60, 1229–1235 (2011).
Fujita, N. et al. Differential Involvement of Atg16L1 in Crohn Disease and Canonical Autophagy: analysis of the organization of the ATG16l1 complex in fibroblasts. J. Biol. Chem. 284, 32602–32609 (2009).
Thachil, E. et al. Abnormal activation of autophagy-induced crinophagy in Paneth cells from patients with Crohn's disease. Gastroenterology 142, 1097–1099. e4 (2012).
Cooney, R. et al. NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nat. Med. 16, 90–97 (2010).
Marchiando, A. M. et al. A deficiency in the autophagy gene Atg16l1 enhances resistance to enteric bacterial infection. Cell Host Microbe 14, 216–224 (2013).
Georges, M. The long and winding road from correlation to causation. Nat. Genet. 43, 180–181 (2011).
Parkes, M. et al. Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn's disease susceptibility. Nat. Genet. 39, 830–832 (2007).
McCarroll, S. A. et al. Deletion polymorphism upstream of IRGM associated with altered IRGM expression and Crohn's disease. Nat. Genet. 40, 1107–1112 (2008).
Van Limbergen, J. et al. Germline variants of IRGM in childhood-onset Crohn's disease. Gut 58, 610–611 (2009).
Brest, P. et al. A synonymous variant in IRGM alters a binding site for miR-196 and causes deregulation of IRGM-dependent xenophagy in Crohn's disease. Nat. Genet. 43, 242–245 (2011).
Yang, S. K. et al. Genome-wide association study of Crohn's disease in Koreans revealed three new susceptibility loci and common attributes of genetic susceptibility across ethnic populations. Gut 63, 80–87 (2014).
Ishibashi, K. et al. Atg16L2, a novel isoform of mammalian Atg16L that is not essential for canonical autophagy despite forming an Atg12-5-16L2 complex. Autophagy 7, 1500–1513 (2011).
Adolph, T. E. et al. Paneth cells as a site of origin for intestinal inflammation. Nature 503, 272–276 (2013).
Kaser, A. et al. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell 134, 743–756 (2008).
Deuring, J. J. et al. Genomic ATG16L1 risk allele-restricted Paneth cell ER stress in quiescent Crohn's disease. Gut http://dx.doi.org/10.1136/gutjnl-2012-303527.
Vandussen, K. L. et al. Genetic variants synthesize to produce Paneth cell phenotypes that define subtypes of Crohn's Disease. Gastroenterology 146, 200–209 (2014).
Cho, J. H. & Brant, S. R. Recent insights into the genetics of inflammatory bowel disease. Gastroenterology 140, 1704–1712 (2011).
D'Haens, G. et al. Challenges to the design, execution, and analysis of randomized controlled trials for inflammatory bowel disease. Gastroenterology 143, 1461–1469 (2012).
Lichtenstein, G. R. et al. Combination of genetic and quantitative serological immune markers are associated with complicated Crohn's disease behavior. Inflamm. Bowel Dis. 17, 2488–2496 (2011).
Willing, B. P. et al. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology 139, 1844–1854 (2010).
Feuerer, M., Hill, J. A., Mathis, D. & Benoist, C. Foxp3+ regulatory T cells: differentiation, specification, subphenotypes. Nat. Immunol. 10, 689–695 (2009).
Hueber, W. et al. Secukinumab, a human anti-IL-17A monoclonal antibody, for moderate to severe Crohn's disease: unexpected results of a randomised, double-blind placebo-controlled trial. Gut 61, 1693–1700 (2012).
Vermeire, S. & Rutgeerts, P. IBD in 2012: Pathogenesis and management of IBD-thinking outside the box. Nat. Rev. Gastroenterol. Hepatol. 10, 67–69 (2013).
Sandborn, W. J. et al. Tofacitinib, an oral Janus kinase inhibitor, in active ulcerative colitis. N. Engl. J. Med. 367, 616–624 (2012).
Sandborn, W. J. et al. Ustekinumab induction and maintenance therapy in refractory Crohn's disease. N. Engl. J. Med. 367, 1519–1528 (2012).
Takahashi, R. et al. SOCS1 is essential for regulatory T cell functions by preventing loss of Foxp3 expression as well as IFN-γ and IL-17A production. J. Exp. Med. 208, 2055–2067 (2011).
Yao, R. et al. MicroRNA-155 modulates TREG and TH17 cells differentiation and TH17 cell function by targeting SOCS1. PLoS ONE 7, e46082 (2012).
Henderson, P. et al. Variation in ICOSLG influences Crohn's disease susceptibility. Gut 60, 1444 (2011).
Marks, D. J. et al. Defective acute inflammation in Crohn's disease: a clinical investigation. Lancet 367, 668–678 (2006).
Segal, A. W. & Loewi, G. Neutrophil dysfunction in Crohn's disease. Lancet 2, 219–221 (1976).
Wu, Y. et al. A chemokine receptor CXCR2 macromolecular complex regulates neutrophil functions in inflammatory Diseases. J. Biol. Chem. 287, 5744–5755 (2012).
Van Assche, G. et al. Progressive multifocal leukoencephalopathy after natalizumab therapy for Crohn's disease. N. Engl. J. Med. 353, 362–368 (2005).
Sandborn, W. J. et al. Vedolizumab as induction and maintenance therapy for Crohn's disease. N. Engl. J. Med. 369, 711–721 (2013).
Feagan, B. G. et al. Vedolizumab as induction and maintenance therapy for ulcerative colitis. N. Engl. J. Med. 369, 699–710 (2013).
Rutgeerts, P. J. et al. A randomised phase I study of etrolizumab (rhuMAb beta7) in moderate to severe ulcerative colitis. Gut 62, 1122–1130 (2013).
Vermeire, S. et al. The mucosal addressin cell adhesion molecule antibody PF-00547659 in ulcerative colitis: a randomised study. Gut 60, 1068–1075 (2011).
Krishnaprasad, K. et al. Inter-observer agreement for Crohn's disease sub-phenotypes using the Montreal Classification: how good are we? A multi-centre Australasian study. J. Crohns Colitis 6, 287–293 (2012).
Sackett, D. L. & Whelan, G. Cancer risk in ulcerative colitis: scientific requirements for the study of prognosis. Gastroenterology 78, 1632–1635 (1980).
Walsh, A. J. & Radford-Smith, G. in IBD—Translating Basic Science into Clinical Practice (eds Targan, S. R., Shanahan, F., Karp, L. C) 212–227 (Wiley-Blackwell, 2010).
Weersma, R. K. et al. Molecular prediction of disease risk and severity in a large Dutch Crohn's disease cohort. Gut 58, 388–395 (2008).
Papa, E. et al. Non-invasive mapping of the gastrointestinal microbiota identifies children with inflammatory bowel disease. PLoS ONE 7, e39242 (2012).
Yang, J. et al. Common SNPs explain a large proportion of the heritability for human height. Nat. Genet. 42, 565–569 (2010).
Janczewska, I. et al. Clinical application of the multigene analysis test in discriminating between ulcerative colitis and Crohn's disease: a retrospective study. Scand. J. Gastroenterol. 47, 162–169 (2012).
Crohn's and Colitis Foundation of Canada Inflammatory Bowel Disease GEM Project. The GEM Project [online], (2014).
Silverberg, M. S., Xu, W., Paterson, A. D. & Croitoru, K. Increased frequency of risk alleles in a cohort of healthy first degree relatives of Crohn's Disease patients—a report from the Michael J. Howorth/CCFC GEM Project. Gastroenterology 140, S268 (2011).
Abreu, M. T. et al. Mutations in NOD2 are associated with fibrostenosing disease in patients with Crohn's disease. Gastroenterology 123, 679–688 (2002).
Brant, S. R. et al. Defining complex contributions of NOD2/CARD15 gene mutations, age at onset, and tobacco use on Crohn's disease phenotypes. Inflamm. Bowel Dis. 9, 281–289 (2003).
Fowler, E. V. et al. ATG16L1 T300A shows strong associations with disease subgroups in a large Australian IBD population: further support for significant disease heterogeneity. Am. J. Gastroenterol. 103, 2519–2526 (2008).
Van Limbergen, J. et al. Autophagy gene ATG16L1 influences susceptibility and disease location but not childhood-onset in Crohn's disease in Northern Europe. Inflamm. Bowel Dis. 14, 338–346 (2008).
Crohn, B. B., Ginzburg, L. & Oppenheimer, G. D. Landmark article Oct 15, 1932. Regional ileitis. A pathological and clinical entity. By Burril B. Crohn, Leon Ginzburg, and Gordon D. Oppenheimer. JAMA 251, 73–79 (1984).
Hancock, L. et al. Clinical and molecular characteristics of isolated colonic Crohn's disease. Inflamm. Bowel Dis. 14, 1667–1677 (2008).
Hume, G. E. et al. Novel NOD2 haplotype strengthens the association between TLR4 Asp299Gly and Crohn's disease in an Australian population. Inflamm. Bowel Dis. 14, 585–590 (2008).
Silverberg, M. S. et al. A population- and family-based study of Canadian families reveals association of HLA DRB1*0103 with colonic involvement in inflammatory bowel disease. Inflamm. Bowel Dis. 9, 1–9 (2003).
Lees, C. W. Characterization of the ∼40,000 patient cohort of the International Inflammatory Bowel Disease Genetics Consortium (IIBDGC). J. Crohns Colitis 7 (Suppl. 1), S4 (2013).
Beaugerie, L., Seksik, P., Nion-Larmurier, I., Gendre, J. P. & Cosnes, J. Predictors of Crohn's disease. Gastroenterology 130, 650–656 (2006).
Lazarev, M. et al. Relationship between proximal Crohn's disease location and disease behavior and surgery: A Cross-Sectional Study of the IBD Genetics Consortium. Am. J. Gastroenterol. 108, 106–112 (2013).
Henckaerts, L. et al. Genetic risk profiling and prediction of disease course in Crohn's disease patients. Clin. Gastroenterol. Hepatol. 7, 972–980. e2 (2009).
Ryan, J. D. et al. Predicting complicated Crohn's disease and surgery: phenotypes, genetics, serology and psychological characteristics of a population-based cohort. Aliment. Pharmacol. Ther. 38, 274–283 (2013).
Dubinsky, M. C. et al. Multidimensional prognostic risk assessment identifies association between IL12B variation and surgery in Crohn's disease. Inflamm. Bowel Dis. 19, 1662–1670 (2013).
Lee, J. C. et al. Human SNP links differential outcomes in inflammatory and infectious disease to a FOXO3-regulated pathway. Cell 155, 57–69 (2013).
Haritunians, T. et al. Genetic predictors of medically refractory ulcerative colitis. Inflamm. Bowel Dis. 16, 1830–1840 (2010).
Radford-Smith, G. et al. Clinical and molecular characterization of medically refractory acute, severe colitis: preliminary results from the International Inflammatory Bowel Disease Genetics Consortium (IIBDGC) Immunochip Study [abstract]. Gastroenterology 144, S470 (2013).
Meier, C. B. et al. Innate immune receptor genetic polymorphisms in pouchitis: is CARD15 a susceptibility factor? Inflamm. Bowel Dis. 11, 965–971 (2005).
Sehgal, R. et al. Genetic risk profiling and gene signature modeling to predict risk of complications after IPAA. Dis. Colon Rectum 55, 239–248 (2012).
Suppiah, V. et al. IL28B is associated with response to chronic hepatitis C interferon-α and ribavirin therapy. Nat. Genet. 41, 1100–1104 (2009).
Torok, H. P., Goke, B. & Konrad, A. Pharmacogenetics of Crohn's disease. Pharmacogenomics 9, 881–893 (2008).
Pierik, M. et al. Tumour necrosis factor-α receptor 1 and 2 polymorphisms in inflammatory bowel disease and their association with response to infliximab. Aliment. Pharmacol. Ther. 20, 303–310 (2004).
Matsukura, H., Ikeda, S., Yoshimura, N., Takazoe, M. & Muramatsu, M. Genetic polymorphisms of tumour necrosis factor receptor superfamily 1A and 1B affect responses to infliximab in Japanese patients with Crohn's disease. Aliment. Pharmacol. Ther. 27, 765–770 (2008).
Steenholdt, C. et al. Genetic polymorphisms of tumour necrosis factor receptor superfamily 1b and fas ligand are associated with clinical efficacy and/or acute severe infusion reactions to infliximab in Crohn's disease. Aliment. Pharmacol. Ther. 36, 650–659 (2012).
Asakura, H. et al. Association of the human lymphocyte-DR2 antigen with Japanese ulcerative colitis. Gastroenterology 82, 413–418 (1982).
Yamazaki, K. et al. Single nucleotide polymorphisms in TNFSF15 confer susceptibility to Crohn's disease. Hum. Mol. Genet. 14, 3499–3506 (2005).
Ng, S. C. et al. Genetics of inflammatory bowel disease in Asia: systematic review and meta-analysis. Inflamm. Bowel Dis. 18, 1164–1176 (2012).
Thia, K. T., Loftus, E. V. Jr, Sandborn, W. J. & Yang, S. K. An update on the epidemiology of inflammatory bowel disease in Asia. Am. J. Gastroenterol. 103, 3167–3182 (2008).
Ng, S. C. et al. Incidence and phenotype of inflammatory bowel disease based on results from the Asia–Pacific Crohn's and colitis epidemiology study. Gastroenterology 145, 158–165 (2013).
Kugathasan, S. et al. Epidemiologic and clinical characteristics of children with newly diagnosed inflammatory bowel disease in Wisconsin: a statewide population-based study. J. Pediatr. 143, 525–531 (2003).
Sakamoto, N. et al. Dietary risk factors for inflammatory bowel disease: a multicenter case-control study in Japan. Inflamm. Bowel Dis. 11, 154–163 (2005).
Koloski, N. A., Bret, L. & Radford-Smith, G. Hygiene hypothesis in inflammatory bowel disease: a critical review of the literature. World J. Gastroenterol. 14, 165–173 (2008).
Chua, K. H. et al. Identification of NOD2/CARD15 mutations in Malaysian patients with Crohn's disease. J. Dig. Dis. 10, 124–130 (2009).
Lv, C. et al. Confirmation of three inflammatory bowel disease susceptibility loci in a Chinese cohort. Int. J. Colorectal Dis. 27, 1465–1472 (2012).
Sugimura, K. et al. A novel NOD2/CARD15 haplotype conferring risk for Crohn disease in Ashkenazi Jews. Am. J. Hum. Genet. 72, 509–518 (2003).
Asano, K. et al. A genome-wide association study identifies three new susceptibility loci for ulcerative colitis in the Japanese population. Nat. Genet. 41, 1325–1329 (2009).
Okada, Y. et al. HLA-Cw*1202-B*5201-DRB1*1502 haplotype increases risk for ulcerative colitis but reduces risk for Crohn's disease. Gastroenterology 141, 864–871. e1–e5 (2011).
Yamazaki, K. et al. A genome-wide association study identifies 2 susceptibility loci for Crohn's disease in a Japanese population. Gastroenterology 144, 781–788 (2013).
Hirano, A. et al. Association study of 71 European Crohn's disease susceptibility loci in a Japanese population. Inflamm. Bowel Dis. 19, 526–533 (2013).
Moon, C. M. et al. Associations between genetic variants in the IRGM gene and inflammatory bowel diseases in the Korean population. Inflamm. Bowel Dis. 19, 106–114 (2013).
Yamazaki, K. et al. Association analysis of genetic variants in IL23R, ATG16L1 and 5p13.1 loci with Crohn's disease in Japanese patients. J. Hum. Genet. 52, 575–583 (2007).
Yang, S. K. et al. Contribution of IL23R but not ATG16L1 to Crohn's disease susceptibility in Koreans. Inflamm. Bowel Dis. 15, 1385–1390 (2009).
Bin, C. et al. Contribution of rs11465788 in IL23R gene to Crohn's disease susceptibility and phenotype in Chinese population. J. Genet. 88, 191–196 (2009).
Yang, S. K. et al. Genome-wide association study of ulcerative colitis in Koreans suggests extensive overlapping of genetic susceptibility with Caucasians. Inflamm. Bowel Dis. 19, 954–966 (2013).
Adeyanju, O. et al. Common NOD2 risk variants in African Americans with Crohn's disease are due exclusively to recent Caucasian admixture. Inflamm. Bowel Dis. 18, 2357–2359 (2012).
Wang, M. H. et al. Contribution of higher risk genes and European admixture to Crohn's disease in African Americans. Inflamm. Bowel Dis. 18, 2277–2287 (2012).
Ventham, N. T., Kennedy, N. A., Nimmo, E. R. & Satsangi, J. Beyond gene discovery in inflammatory bowel disease: the emerging role of epigenetics. Gastroenterology 145, 293–308 (2013).
Nguyen, H. T. et al. Crohn's disease-associated adherent invasive Escherichia coli modulate levels of micrornas in intestinal epithelial cells to reduce autophagy. Gastroenterology 146, 508–519 (2013).
Lu, C. et al. MIR106B and MIR93 prevent removal of bacteria from epithelial cells by disrupting ATG16L1-mediated autophagy. Gastroenterology 146, 188–199 (2014).
Trynka, G. et al. Chromatin marks identify critical cell types for fine mapping complex trait variants. Nat. Genet. 45, 124–130 (2013).
Sato, T. et al. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature 469, 415–418 (2010).
Conway, K. L. et al. Atg16l1 is required for autophagy in intestinal epithelial cells and protection of mice from Salmonella infection. Gastroenterology 145, 1347–1357 (2013).
Yilmaz, O. H. et al. mTORC1 in the Paneth cell niche couples intestinal stem-cell function to calorie intake. Nature 486, 490–495 (2012).
Acknowledgements
J.V.L. acknowledges support from the North American Society for Paediatric Gastroenterology, Hepatology and Nutrition Foundation and Crohn's & Colitis Foundation of America Young Investigator Development Award 2013.
Author information
Authors and Affiliations
Contributions
All authors made equal contributions to all aspects of this manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Table 1
Prioritized candidate genes on IBD susceptibility loci. (DOC 40 kb)
Rights and permissions
About this article
Cite this article
Van Limbergen, J., Radford-Smith, G. & Satsangi, J. Advances in IBD genetics. Nat Rev Gastroenterol Hepatol 11, 372–385 (2014). https://doi.org/10.1038/nrgastro.2014.27
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrgastro.2014.27
This article is cited by
-
The role of sphingosine-1-phosphate in autophagy and related disorders
Cell Death Discovery (2023)
-
Mutation spectrum of NOD2 reveals recessive inheritance as a main driver of Early Onset Crohn’s Disease
Scientific Reports (2021)
-
Epithelial production of elastase is increased in inflammatory bowel disease and causes mucosal inflammation
Mucosal Immunology (2021)
-
Macrophage-derived EDA-A2 inhibits intestinal stem cells by targeting miR-494/EDA2R/β-catenin signaling in mice
Communications Biology (2021)
-
Paediatric Inflammatory Bowel Disease and its Relationship with the Microbiome
Microbial Ecology (2021)