Key Points
-
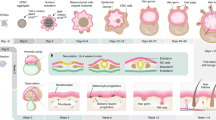
By combining genetic engineering and cell-biological studies, skin biologists are discovering the mechanisms that underlie the development and differentiation of the epidermis and hair follicles of the skin.
-
Populations of stem cells exist in the basal layer of the epidermis and the hair-follicle bulge. The Notch and Wnt pathways are involved in the decision of the progeny of these cells to commit to an epidermal or hair-follicle fate.
-
Gene-targeting studies of epidermal transcription factor genes have revealed important functional information about the involvement of these factors in balancing differentiation with proliferation and about their role in human genetic skin disorders.
-
Altering the balance between cell proliferation and cell adhesion is required for wound healing and occurs aberrantly in certain skin cancers and skin disorders. New insights into such disorders are being provided by gene targeting and cell-biological studies of the components that make up the main cell junctions of the epidermis.
-
Despite recent insights into the genetic basis of human skin disorders, the therapeutic options for the autosomal-dominant disorders remain bleak. But gene therapy has been successfully used to restore keratinocyte function in certain recessive skin disorders.
-
Recent genetic and cell-biological studies have highlighted the importance of cell adhesion, basement membrane assembly and tissue architecture in contributing to the proper balance and spatial arrangement of epidermal growth and differentiation. But many questions about skin biology remain, which new tools and model systems should help to resolve.
Abstract
At the surface of the skin, the epidermis serves as the armour for the body. Scientists are now closer than ever to understanding how the epidermis accomplishes this extraordinary feat, and is able to survive and replenish itself under the harshest conditions that face any tissue. By combining genetic engineering with cell-biological studies and with human genome data analyses, skin biologists are discovering the mechanisms that underlie the development and differentiation of the epidermis and hair follicles of the skin. This explosion of knowledge paves the way for new discoveries into the genetic bases of human skin disorders and for developing new therapeutics.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Rochat, A., Kobayashi, K. & Barrandon, Y. Location of stem cells of human hair follicles by clonal analysis. Cell 76, 1063–1073 (1994).
Watt, F. M. & Hogan, B. L. Out of Eden: stem cells and their niches. Science 287, 1427–1430 (2000).
Oshima, H., Rochat, A., Kedzia, C., Kobayashi, K. & Barrandon, Y. Morphogenesis and renewal of hair follicles from adult multipotent stem cells. Cell 104, 233–245 (2001).
Taylor, G., Lehrer, M. S., Jensen, P. J., Sun, T. T. & Lavker, R. M. Involvement of follicular stem cells in forming not only the follicle but also the epidermis. Cell 102, 451–461 (2000).References 3 and 4 describe the location and characteristics of epidermal stem cells that reside in the bulge compartment of hair follicles.
Jones, P. H. & Watt, F. M. Separation of human epidermal stem cells from transit amplifying cells on the basis of differences in integrin function and expression. Cell 73, 713–724 (1993).
Jones, P. H., Harper, S. & Watt, F. M. Stem cell patterning and fate in human epidermis. Cell 80, 83–93 (1995).
Barrandon, Y. & Green, H. Three clonal types of keratinocyte with different capacities for multiplication. Proc. Natl Acad. Sci. USA 84, 2302–2306 (1987).
Watt, F. M. Epidermal stem cells: markers, patterning and the control of stem cell fate. Phil. Trans. R. Soc. Lond. B 353, 831–837 (1998).
Tani, H., Morris, R. J. & Kaur, P. Enrichment for murine keratinocyte stem cells based on cell surface phenotype. Proc. Natl Acad. Sci. USA 97, 10960–10965 (2000).
Pellegrini, G. et al. p63 identifies keratinocyte stem cells. Proc. Natl Acad. Sci. USA 98, 3156–3161 (2001).
Yang, A. et al. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature 398, 714–718 (1999).
Mills, A. A. et al. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature 398, 708–713 (1999).References 11 and 12 describe the identification and cloning of Trp63 and the characterization of mice that lack p63 function. These papers show the importance of p63 in epidermal morphogenesis and its role in controlling proliferation versus differentiation.
Waikel, R. L., Kawachi, Y., Waikel, P. A., Wang, X. J. & Roop, D. R. Deregulated expression of c-Myc depletes epidermal stem cells. Nature Genet. 28, 165–168 (2001).
Arnold, I. & Watt, F. M. c-Myc activation in transgenic mouse epidermis results in mobilization of stem cells and differentiation of their progeny. Curr. Biol. 11, 558–568 (2001).
Bull, J. J. et al. Contrasting localization of c-Myc with other Myc superfamily transcription factors in the human hair follicle and during the hair growth cycle. J. Invest. Dermatol. 116, 617–622 (2001).
Barker, N. & Clevers, H. Catenins, Wnt signaling and cancer. Bioessays 22, 961–965 (2000).
Millar, S. E. et al. WNT signaling in the control of hair growth and structure. Dev. Biol. 207, 133–149 (1999).
St-Jacques, B. et al. Sonic hedgehog signaling is essential for hair development. Curr. Biol. 8, 1058–1068 (1998).
DasGupta, R. & Fuchs, E. Multiple roles for activated LEF/TCF transcription complexes during hair follicle development and differentiation. Development 126, 4557–4568 (1999).
Gat, U., DasGupta, R., Degenstein, L. & Fuchs, E. De novo hair follicle morphogenesis and hair tumors in mice expressing a truncated β-catenin in skin. Cell 95, 605–614 (1998).This paper showed that ectopic β-catenin signalling in the postnatal epidermis was sufficient to induce hair-follicle differentiation pathways in cells that would otherwise remain epidermal in nature.
Merrill, B. J., Gat, U., DasGupta, R. & Fuchs, E. Tcf3 and Lef1 regulate lineage differentiation of multipotent stem cells in skin. Genes Dev. 15, 1688–1705 (2001).
Huelsken, J., Vogel, R., Erdmann, B., Cotsarelis, G. & Birchmeier, W. β-Catenin controls hair follicle morphogenesis and stem cell differentiation in the skin. Cell 105, 533–545 (2001).
Niemann, C., Owens, D. M., Hulsken, J., Birchmeier, W. & Watt, F. M. Expression of ΔNLef1 in mouse epidermis results in differentiation of hair follicles into squamous epidermal cysts and formation of skin tumours. Development 129, 95–109 (2002).
Zhou, P., Byrne, C., Jacobs, J. & Fuchs, E. Lymphoid enhancer factor 1 directs hair follicle patterning and epithelial cell fate. Genes Dev. 9, 700–713 (1995).
Lewis, J. Notch signalling and the control of cell fate choices in vertebrates. Semin. Cell Dev. Biol. 9, 583–589 (1998).
Morrison, S. J. et al. Transient Notch activation initiates an irreversible switch from neurogenesis to gliogenesis by neural crest stem cells. Cell 101, 499–510 (2000).
Artavanis-Tsakonas, S., Rand, M. D. & Lake, R. J. Notch signaling: cell fate control and signal integration in development. Science 284, 770–776 (1999).
Powell, B. C., Passmore, E. A., Nesci, A. & Dunn, S. M. The Notch signalling pathway in hair growth. Mech. Dev. 78, 189–192 (1998).
Favier, B. et al. Localisation of members of the notch system and the differentiation of vibrissa hair follicles: receptors, ligands, and fringe modulators. Dev. Dyn. 218, 426–437 (2000).
Lin, M. H., Leimeister, C., Gessler, M. & Kopan, R. Activation of the Notch pathway in the hair cortex leads to aberrant differentiation of the adjacent hair-shaft layers. Development 127, 2421–2432 (2000).
Lowell, S., Jones, P., Le Roux, I., Dunne, J. & Watt, F. M. Stimulation of human epidermal differentiation by delta–notch signalling at the boundaries of stem-cell clusters. Curr. Biol. 10, 491–500 (2000).
Sasaki, Y. et al. The p53 family member genes are involved in the Notch signal pathway. J. Biol. Chem. 277, 719–724 (2002).
Rangarajan, A. et al. Notch signaling is a direct determinant of keratinocyte growth arrest and entry into differentiation. EMBO J. 20, 3427–3436 (2001).This paper describes the conditional, inducible ablation of Notch1 in keratinocytes. It underscores the importance of Notch1 signalling in regulating epidermal differentiation.
Xia, X. et al. Loss of presenilin 1 is associated with enhanced β-catenin signaling and skin tumorigenesis. Proc. Natl Acad. Sci. USA 11, 10863–10868 (2001).
Killick, R. et al. Presenilin 1 independently regulates β-catenin stability and transcriptional activity. J. Biol. Chem. 276, 48554–48561 (2001).
Palacino, J. J. et al. Presenilin 1 regulates β-catenin-mediated transcription in a glycogen synthase kinase-3-independent fashion. J. Biol. Chem. 276, 38563–38569 (2001).
Soriano, S. et al. Presenilin 1 negatively regulates β-catenin/T cell factor/lymphoid enhancer factor-1 signaling independently of β-amyloid precursor protein and notch processing. J. Cell Biol. 152, 785–794 (2001).
Ross, D. A. & Kadesch, T. The notch intracellular domain can function as a coactivator for lef-1. Mol. Cell. Biol. 21, 7537–7544 (2001).
Fuchs, E., Merrill, B. J., Jamora, C. & DasGupta, R. At the roots of a never-ending cycle. Dev. Cell 1, 13–25 (2001).
Altmann, C. R. & Brivanlou, A. H. Neural patterning in the vertebrate embryo. Int. Rev. Cytol. 203, 447–482 (2001).
Wilson, S. I. et al. The status of Wnt signalling regulates neural and epidermal fates in the chick embryo. Nature 411, 325–330 (2001).
Tseng, H., Biegel, J. A. & Brown, R. S. Basonuclin is associated with the ribosomal RNA genes on human keratinocyte mitotic chromosomes. J. Cell Sci. 112, 3039–3047 (1999).
Maytin, E. V. et al. Keratin 10 gene expression during differentiation of mouse epidermis requires transcription factors C/EBP and AP-2. Dev. Biol. 216, 164–181 (1999).
Faus, I., Hsu, H. J. & Fuchs, E. Oct-6: a regulator of keratinocyte gene expression in stratified squamous epithelia. Mol. Cell. Biol. 14, 3263–3275 (1994).
Andersen, B. et al. Functions of the POU domain genes Skn-1a/i and Tst-1/Oct-6/SCIP in epidermal differentiation. Genes Dev. 11, 1873–1884 (1997).
Oettgen, P. et al. Characterization of ESE-2, a novel ESE-1-related Ets transcription factor that is restricted to glandular epithelium and differentiated keratinocytes. J. Biol. Chem. 274, 29439–29452 (1999).
Segre, J. A., Bauer, C. & Fuchs, E. Klf4 is a transcription factor required for establishing the barrier function of the skin. Nature Genet. 22, 356–360 (1999).
Li, M. et al. RXR-α ablation in skin keratinocytes results in alopecia and epidermal alterations. Development 128, 675–688 (2001).
Li, Q. et al. IKK1-deficient mice exhibit abnormal development of skin and skeleton. Genes Dev. 13, 1322–1328 (1999).
Hu, Y. et al. Abnormal morphogenesis but intact IKK activation in mice lacking the IKKα subunit of IκB kinase. Science 284, 316–320 (1999).
Takeda, K. et al. Limb and skin abnormalities in mice lacking IKKα. Science 284, 313–316 (1999).
Hu, Y. et al. IKKα controls formation of the epidermis independently of NF-κB. Nature 410, 710–714 (2001).This paper showed that IKKα is involved in the production of a secreted factor that is crucial for terminal differentiation of mouse keratinocytes.
Makris, C. et al. Female mice heterozygous for IKKγ/NEMO deficiencies develop a dermatopathy similar to the human X-linked disorder incontinentia pigmenti. Mol. Cell 5, 969–979 (2000).
Schmidt-Supprian, M. et al. NEMO/IKKγ-deficient mice model incontinentia pigmenti. Mol. Cell 5, 981–992 (2000).
The International Incontinentia Pigmenti (IP) Consortium. Genomic rearrangement in NEMO impairs NF-κB activation and is a cause of incontinentia pigmenti. Nature 405, 466–472 (2000).
Seitz, C. S., Lin, Q., Deng, H. & Khavari, P. A. Alterations in NF-κB function in transgenic epithelial tissue demonstrate a growth inhibitory role for NF-κB. Proc. Natl Acad. Sci. USA 95, 2307–2312 (1998).
Fuchs, E. & Green, H. Regulation of terminal differentiation of cultured human keratinocytes by vitamin A. Cell 25, 617–625 (1981).
Kopan, R., Traska, G. & Fuchs, E. Retinoids as important regulators of terminal differentiation: examining keratin expression in individual epidermal cells at various stages of keratinization. J. Cell Biol. 105, 427–440 (1987).
Dolle, P. et al. Differential expression of genes encoding α, β and γ retinoic acid receptors and CRABP in the developing limbs of the mouse. Nature 342, 702–705 (1989).
Xiao, J. H., Durand, B., Chambon, P. & Voorhees, J. J. Endogenous retinoic acid receptor (RAR)–retinoid X receptor (RXR) heterodimers are the major functional forms regulating retinoid-responsive elements in adult human keratinocytes. Binding of ligands to RAR only is sufficient for RAR–RXR heterodimers to confer ligand-dependent activation of hRARβ2/RARE (DR5). J. Biol. Chem. 270, 3001–3011 (1995).
Imakado, S. et al. Targeting expression of a dominant-negative retinoic acid receptor mutant in the epidermis of transgenic mice results in loss of barrier function. Genes Dev. 9, 317–329 (1995).
Saitou, M. et al. Inhibition of skin development by targeted expression of a dominant-negative retinoic acid receptor. Nature 374, 159–162 (1995).
Ahmad, W., Panteleyev, A. A., Sundberg, J. P. & Christiano, A. M. Molecular basis for the rhino (hrrh-8J) phenotype: a nonsense mutation in the mouse hairless gene. Genomics 53, 383–386 (1998).
Leask, A., Rosenberg, M., Vassar, R. & Fuchs, E. Regulation of a human epidermal keratin gene: sequences and nuclear factors involved in keratinocyte-specific transcription. Genes Dev. 4, 1985–1998 (1990).
Snape, A. M., Jonas, E. A. & Sargent, T. D. KTF-1, a transcriptional activator of Xenopus embryonic keratin expression. Development 109, 157–165 (1990).
Leask, A., Byrne, C. & Fuchs, E. Transcription factor AP2 and its role in epidermal-specific gene expression. Proc. Natl Acad. Sci. USA 88, 7948–7952 (1991).
Byrne, C., Tainsky, M. & Fuchs, E. Programming gene expression in developing epidermis. Development 120, 2369–2383 (1994).
Casatorres, J., Navarro, J. M., Blessing, M. & Jorcano, J. L. Analysis of the control of expression and tissue specificity of the keratin 5 gene, characteristic of basal keratinocytes. Fundamental role of an AP-1 element. J. Biol. Chem. 269, 20489–20496 (1994).
DiSepio, D. et al. The proximal promoter of the mouse loricrin gene contains a functional AP-1 element and directs keratinocyte-specific but not differentiation-specific expression. J. Biol. Chem. 270, 10792–10799 (1995).
Jang, S. I., Steinert, P. M. & Markova, N. G. Activator protein 1 activity is involved in the regulation of the cell type-specific expression from the proximal promoter of the human profilaggrin gene. J. Biol. Chem. 271, 24105–24114 (1996).
LaPres, J. J. & Hudson, L. G. Identification of a functional determinant of differentiation-dependent expression in the involucrin gene. J. Biol. Chem. 271, 23154–23160 (1996).
Byrne, C. Regulation of gene expression in developing epidermal epithelia. Bioessays 19, 691–698 (1997).
Sinha, S., Degenstein, L., Copenhaver, C. & Fuchs, E. Defining the regulatory factors required for epidermal gene expression. Mol. Cell. Biol. 20, 2543–2555 (2000).
Sinha, S. & Fuchs, E. Identification and dissection of an enhancer controlling epithelial gene expression in skin. Proc. Natl Acad. Sci. USA 98, 2455–2460 (2001).
Tomic-Canic, M., Komine, M., Freedberg, I. M. & Blumenberg, M. Epidermal signal transduction and transcription factor activation in activated keratinocytes. J. Dermatol. Sci. 17, 167–181 (1998).
Efimova, T., LaCelle, P., Welter, J. F. & Eckert, R. L. Regulation of human involucrin promoter activity by a protein kinase C, Ras, MEKK1, MEK3, p38/RK, AP1 signal transduction pathway. J. Biol. Chem. 273, 24387–24395 (1998).
Jochum, W., Passegue, E. & Wagner, E. F. AP-1 in mouse development and tumorigenesis. Oncogene 20, 2401–2412 (2001).
Hilberg, F., Aguzzi, A., Howells, N. & Wagner, E. F. c-Jun is essential for normal mouse development and hepatogenesis. Nature 365, 179–181 (1993).
Gruda, M. C. et al. Expression of FosB during mouse development: normal development of FosB knockout mice. Oncogene 12, 2177–2185 (1996).
Zhang, J. et al. Neural tube, skeletal and body wall defects in mice lacking transcription factor AP-2. Nature 381, 238–241 (1996).
Eferl, R. et al. Functions of c-Jun in liver and heart development. J. Cell Biol. 145, 1049–1061 (1999).
Passegue, E., Jochum, W., Behrens, A., Ricci, R. & Wagner, E. F. JunB can substitute for Jun in mouse development and cell proliferation. Nature Genet. 2002 Jan 2; [epub ahead of print].
Green, K. J. & Gaudry, C. A. Are desmosomes more than tethers for intermediate filaments? Nature Rev. Mol. Cell Biol. 1, 208–216 (2000).
Vasioukhin, V., Bauer, C., Yin, M. & Fuchs, E. Directed actin polymerization is the driving force for epithelial cell–cell adhesion. Cell 100, 209–219 (2000).
Runswick, S. K., O'Hare, M. J., Jones, L., Streuli, C. H. & Garrod, D. R. Desmosomal adhesion regulates epithelial morphogenesis and cell positioning. Nature Cell Biol. 3, 823–830 (2001).
Peifer, M. & Polakis, P. Wnt signaling in oncogenesis and embryogenesis — a look outside the nucleus. Science 287, 1606–1609 (2000).
Powell, S. M. et al. APC mutations occur early during colorectal tumorigenesis. Nature 359, 235–237 (1992).
Chan, E. F., Gat, U., McNiff, J. M. & Fuchs, E. A common human skin tumour is caused by activating mutations in β-catenin. Nature Genet. 21, 410–413 (1999).References 87 and 88 show the role of activating mutations in β-catenin and of members in the signalling cascade that are involved in the degradation of β-catenin, in the generation of tumours.
Shiozaki, H. et al. Immunohistochemical detection of α-catenin expression in human cancers. Am. J. Pathol. 144, 667–674 (1994).
Schipper, J. H. et al. Expression of E-cadherin in skin carcinomas. J. Dermatol. 23, 104–110 (1996).
Adams, C. L. & Nelson, W. J. Cytomechanics of cadherin-mediated cell–cell adhesion. Curr. Opin. Cell Biol. 10, 572–577 (1998).
Yap, A. S., Niessen, C. M. & Gumbiner, B. M. The juxtamembrane region of the cadherin cytoplasmic tail supports lateral clustering, adhesive strengthening, and interaction with p120ctn. J. Cell Biol. 141, 779–789 (1998).
Vasioukhin, V., Bauer, C., Degenstein, L., Wise, B. & Fuchs, E. Hyperproliferation and defects in epithelial polarity upon conditional ablation of α-catenin in skin. Cell 104, 605–617 (2001).
McGrath, J. A. et al. Mutations in the plakophilin 1 gene result in ectodermal dysplasia/skin fragility syndrome. Nature Genet. 17, 240–244 (1997).
Armstrong, D. K. et al. Haploinsufficiency of desmoplakin causes a striate subtype of palmoplantar keratoderma. Hum. Mol. Genet. 8, 143–148 (1999).
DiPersio, C. M., Hodivala-Dilke, K. M., Jaenisch, R., Kreidberg, J. A. & Hynes, R. O. α3β1-Integrin is required for normal development of the epidermal basement membrane. J. Cell Biol. 137, 729–742 (1997).
Brakebusch, C. et al. Skin and hair follicle integrity is crucially dependent on β1 integrin expression on keratinocytes. EMBO J. 19, 3990–4003 (2000).
Raghavan, S., Bauer, C., Mundschau, G., Li, Q. & Fuchs, E. Conditional ablation of β1 integrin in skin. Severe defects in epidermal proliferation, basement membrane formation, and hair follicle invagination. J. Cell Biol. 150, 1149–1160 (2000).
Dowling, J., Yu, Q. C. & Fuchs, E. β4 integrin is required for hemidesmosome formation, cell adhesion and cell survival. J. Cell Biol. 134, 559–572 (1996).
Georges-Labouesse, E. et al. Absence of integrin α6 leads to epidermolysis bullosa and neonatal death in mice. Nature Genet. 13, 370–373 (1996).
Van der Neut, R., Krimpenfort, P., Calafat, J., Niessen, C. M. & Sonnenberg, A. Epithelial detachment due to absence of hemidesmosomes in integrin β4 null mice. Nature Genet. 13, 366–369 (1996).
Borradori, L. & Sonnenberg, A. Structure and function of hemidesmosomes: more than simple adhesion complexes. J. Invest. Dermatol. 112, 411–418 (1999).
Christiano, A. M. & Uitto, J. Molecular complexity of the cutaneous basement membrane zone. Revelations from the paradigms of epidermolysis bullosa. Exp. Dermatol. 5, 1–11 (1996).
Guo, L., Degenstein, L. & Fuchs, E. Keratinocyte growth factor is required for hair development but not for wound healing. Genes Dev. 10, 165–175 (1996).
Andra, K. et al. Targeted inactivation of plectin reveals essential function in maintaining the integrity of skin, muscle, and heart cytoarchitecture. Genes Dev. 11, 3143–3156 (1997).
Khavari, P. A. Gene therapy for genetic skin disease. J. Invest. Dermatol. 110, 462–467 (1998).This paper reviews the scope of gene therapy in treating debilitating human skin disorders.
Hansen, L. A. et al. The epidermal growth factor receptor is required to maintain the proliferative population in the basal compartment of epidermal tumors. Cancer Res. 60, 3328–3332 (2000).
DiPersio, C. M. et al. α3β1 and α6β4 integrin receptors for laminin-5 are not essential for epidermal morphogenesis and homeostasis during skin development. J. Cell Sci. 113, 3051–3062 (2000).
Szabowski, A. et al. c-Jun and JunB antagonistically control cytokine-regulated mesenchymal–epidermal interaction in skin. Cell 103, 745–755 (2000).
Huffman, J. A., Hull, W. M., Dranoff, G., Mulligan, R. C. & Whitsett, J. A. Pulmonary epithelial cell expression of GM-CSF corrects the alveolar proteinosis in GM-CSF-deficient mice. J. Clin. Invest. 97, 649–655 (1996).
Metcalf, D. et al. The biological consequences of excess GM-CSF levels in transgenic mice also lacking high-affinity receptors for GM-CSF. Leukemia 12, 353–362 (1998).
Reynolds, A. J. & Jahoda, C. A. Hair matrix germinative epidermal cells confer follicle-inducing capabilities on dermal sheath and high passage papilla cells. Development 122, 3085–3094 (1996).
Suzuki, K. et al. Defective terminal differentiation and hypoplasia of the epidermis in mice lacking the Fgf10 gene. FEBS Lett. 481, 53–56 (2000).
Luetteke, N. C. et al. The mouse waved-2 phenotype results from a point mutation in the EGF receptor tyrosine kinase. Genes Dev. 8, 399–413 (1994).
Miettinen, P. J. et al. Epithelial immaturity and multiorgan failure in mice lacking epidermal growth factor receptor. Nature 376, 337–341 (1995).
Sibilia, M. & Wagner, E. F. Strain-dependent epithelial defects in mice lacking the EGF receptor. Science 269, 234–238 (1995).
Threadgill, D. W. et al. Targeted disruption of mouse EGF receptor: effect of genetic background on mutant phenotype. Science 269, 230–234 (1995).
Hansen, L. A. et al. Genetically null mice reveal a central role for epidermal growth factor receptor in the differentiation of the hair follicle and normal hair development. Am. J. Pathol. 150, 1959–1975 (1997).
Bailleul, B. et al. Skin hyperkeratosis and papilloma formation in transgenic mice expressing a ras oncogene from a suprabasal keratin promoter. Cell 62, 697–708 (1990).
Wang, X. J., Greenhalgh, D. A., Eckhardt, J. N., Rothnagel, J. A. & Roop, D. R. Epidermal expression of transforming growth factor-α in transgenic mice: induction of spontaneous and 12-O-tetradecanoylphorbol-13-acetate-induced papillomas via a mechanism independent of Ha-ras activation or overexpression. Mol. Carcinog. 10, 15–22 (1994).
Woodworth, C. D., Gaiotti, D., Michael, E., Hansen, L. & Nees, M. Targeted disruption of the epidermal growth factor receptor inhibits development of papillomas and carcinomas from human papillomavirus-immortalized keratinocytes. Cancer Res. 60, 4397–4402 (2000).
Sibilia, M. et al. The EGF receptor provides an essential survival signal for SOS-dependent skin tumor development. Cell 102, 211–220 (2000).
Blumberg, H. et al. Interleukin 20: discovery, receptor identification, and role in epidermal function. Cell 104, 9–19 (2001).
Sano, S. et al. Keratinocyte-specific ablation of Stat3 exhibits impaired skin remodeling, but does not affect skin morphogenesis. EMBO J. 18, 4657–4668 (1999).
Vasioukhin, V. & Fuchs, E. Actin dynamics and cell–cell adhesion in epithelia. Curr. Opin. Cell Biol. 13, 76–84 (2001).
Acknowledgements
We thank R. DasGupta, B. J. Merrill and C. Jamora for their critical reading of this review and for their valuable suggestions.
Author information
Authors and Affiliations
Corresponding author
Related links
Glossary
- MULTIPOTENT STEM CELL
-
A stem cell that has the potential to give rise to multiple cell lineages.
- KERATIN
-
A cytoskeletal filament that is typically 10 nm in diameter.
- SEBOCYTE
-
A cell of the sebaceous gland.
- NEURAL INDUCTION
-
The specification of cells that give rise to the neural tube and, ultimately, to the central nervous system.
- EPIBLAST CELL
-
A cell in the early embryo that gives rise to all three definitive germ layers of the embryo: the ectoderm, mesoderm and endoderm.
- CORNIFIED ENVELOPE
-
An extremely tough protein lipid polymer structure that forms just below the cytoplasmic membrane of cells of the cornified layer.
- E-CADHERIN
-
A homophilic cell-adhesion molecule that is an important component of the adherens junctions.
- ACTIN CYTOSKELETON
-
A microfilament network that consists of filaments that are 6 nm in diameter and made up of polymerized actin. The actin cytoskeleton forms the main component of the cellular contractile machinery.
- INTERMEDIATE FILAMENT CYTOSKELETON
-
A network that consists of filaments, typically 10 nm in diameter, that contributes to the mechanical strength of cells.
- INTEGRIN
-
A transmembrane protein that functions as a heterodimer and is involved in cell–cell and cell–extracellular-matrix interactions.
- REVERSE GENETICS
-
A genetic analysis that proceeds from genotype to phenotype by gene-manipulation techniques, such as homologous recombination in embryonic stem cells.
Rights and permissions
About this article
Cite this article
Fuchs, E., Raghavan, S. Getting under the skin of epidermal morphogenesis. Nat Rev Genet 3, 199–209 (2002). https://doi.org/10.1038/nrg758
Issue Date:
DOI: https://doi.org/10.1038/nrg758
This article is cited by
-
Current status of skin cancers with a focus on immunology and immunotherapy
Cancer Cell International (2023)
-
β-endorphin suppresses ultraviolet B irradiation-induced epidermal barrier damage by regulating inflammation-dependent mTORC1 signaling
Scientific Reports (2023)
-
THY1-mediated mechanisms converge to drive YAP activation in skin homeostasis and repair
Nature Cell Biology (2022)
-
Gene duplications and gene loss in the epidermal differentiation complex during the evolutionary land-to-water transition of cetaceans
Scientific Reports (2021)
-
Proteomic characterization of HaCaT keratinocytes provides new insights into changes associated with SDS exposure
Biomedical Dermatology (2020)