Key Points
-
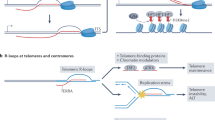
R loops consist of RNA–DNA hybrids that accumulate at preferred regions all over the genome, such as pericentromeric DNA, telomeres, ribosomal DNA or transcription termination regions.
-
R loops are an important source of replication stress and genome instability, which are hallmarks of cancer. As such, R loops seem to be a cause of tumorigenesis.
-
Cells have two ways to limit the number of R loops in the genome: mechanisms that remove R loops, such as ribonucleases and RNA–DNA helicases; and mechanisms that prevent R-loop accumulation, such as RNA-processing factors or topoisomerase 1.
-
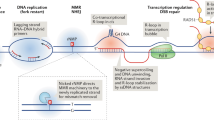
R loops may play a part in transcription activation by directly affecting the chromatin structure of promoters and the recruitment of transcription or chromatin-remodelling factors. Such R loops may be formed by antisense non-coding RNAs.
-
R loops trigger chromatin condensation and heterochromatin formation, which may explain their ability to repress or silence gene expression and to stall replication-fork progression, putatively leading to replication-fork breakage as the main source of R-loop-mediated genome instability.
-
BRCA1 and BRCA2 have functions in double-strand break repair and/or replication-fork protection that may contribute to resolving intermediate structures such as stalled or broken replication forks that are generated as a consequence of R loops — thus these proteins facilitate the removal of R loops.
Abstract
R loops are nucleic acid structures composed of an RNA–DNA hybrid and a displaced single-stranded DNA. Recently, evidence has emerged that R loops occur more often in the genome and have greater physiological relevance, including roles in transcription and chromatin structure, than was previously predicted. Importantly, however, R loops are also a major threat to genome stability. For this reason, several DNA and RNA metabolism factors prevent R-loop formation in cells. Dysfunction of these factors causes R-loop accumulation, which leads to replication stress, genome instability, chromatin alterations or gene silencing, phenomena that are frequently associated with cancer and a number of genetic diseases. We review the current knowledge of the mechanisms controlling R loops and their putative relationship with disease.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Roberts, R. W. & Crothers, D. M. Stability and properties of double and triple helices: dramatic effects of RNA or DNA backbone composition. Science 258, 1463–1466 (1992).
Roy, D., Zhang, Z., Lu, Z., Hsieh, C. L. & Lieber, M. R. Competition between the RNA transcript and the nontemplate DNA strand during R-loop formation in vitro: a nick can serve as a strong R-loop initiation site. Mol. Cell. Biol. 30, 146–159 (2010).
Roy, D. & Lieber, M. R. G clustering is important for the initiation of transcription-induced R-loops in vitro, whereas high G density without clustering is sufficient thereafter. Mol. Cell. Biol. 29, 3124–3133 (2009).
Duquette, M. L., Handa, P., Vincent, J. A., Taylor, A. F. & Maizels, N. Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes Dev. 18, 1618–1629 (2004).
Aguilera, A. & Garcia-Muse, T. R loops: from transcription byproducts to threats to genome stability. Mol. Cell 46, 115–124 (2012).
Aguilera, A. The connection between transcription and genomic instability. EMBO J. 21, 195–201 (2002).
Huertas, P. & Aguilera, A. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol. Cell 12, 711–721 (2003). This paper provides the first demonstration that R loops cause genome instability and that mRNP biogenesis factors prevent R-loop formation. It shows that R loops accumulate in yeast mutants lacking the Hpr1 subunit of the THO complex and that hyper-recombination in these mutants is partially dependent on the nascent mRNA and R-loop accumulation.
Li, X. & Manley, J. L. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell 122, 365–378 (2005). This is the first evidence in vertebrate cells that depletion of a splicing factor causes genome instability in the form of chromosomal rearrangements and mutagenesis mediated by R loops, as shown for chicken DT40 and human HeLa cells depleted of the SRSF1 protein.
Skourti-Stathaki, K., Proudfoot, N. J. & Gromak, N. Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol. Cell 42, 794–805 (2011). This paper connects RNA:DNA helicase SETX deficiency with R-loop accumulation in human cells. The authors propose that R loops form at G-rich termination pausing sites and are resolved by SETX, which would promote 3′ mRNA degradation by XRN2 and transcription termination.
Castellano-Pozo, M. et al. R loops are linked to histone H3 S10 phosphorylation and chromatin condensation. Mol. Cell 52, 583–590 (2013). This work shows a functional link between R loops and the H3S10P chromatin condensation mark. Yeast, worm and human cells depleted of THO subunits show increased levels of H3S10P, which is suppressed by RNase H1 overexpression. Enrichment of the H3K9me2 heterochromatic mark is also shown in C. elegans . It is proposed that chromatin condensation linked to R loops is a strong barrier to replication progression as a major source of replication stress and genome instability.
Wongsurawat, T., Jenjaroenpun, P., Kwoh, C. K. & Kuznetsov, V. Quantitative model of R-loop forming structures reveals a novel level of RNA-DNA interactome complexity. Nucleic Acids Res. 40, e16 (2012).
Ginno, P. A., Lim, Y. W., Lott, P. L., Korf, I. & Chedin, F. GC skew at the 5′ and 3 ends of human genes links R-loop formation to epigenetic regulation and transcription termination. Genome Res. 23, 1590–1600 (2013).
Ginno, P. A., Lott, P. L., Christensen, H. C., Korf, I. & Chedin, F. R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol. Cell 45, 814–825 (2012). This is the first genome-wide analysis of R-loop locations. It shows that unmethylated human CpG island promoters are characterized by a positive GC skew and by the formation of R loops, which protect from de novo DNA methylation.
Chan, Y. A. et al. Genome-wide profiling of yeast DNA:RNA hybrid prone sites with DRIP-chip. PLoS Genet. 10, e1004288 (2014).
El Hage, A., Webb, S., Kerr, A. & Tollervey, D. Genome-wide distribution of RNA-DNA hybrids identifies RNase H targets in tRNA genes, retrotransposons and mitochondria. PLoS Genet. 10, e1004716 (2014).
Skourti-Stathaki, K. & Proudfoot, N. J. A double-edged sword: R loops as threats to genome integrity and powerful regulators of gene expression. Genes Dev. 28, 1384–1396 (2014).
Sollier, J. & Cimprich, K. A. Breaking bad: R-loops and genome integrity. Trends Cell Biol. http://dx.doi.org/10.1016/j.tcb.2015.05.003 (2015).
Cerritelli, S. M. & Crouch, R. J. Ribonuclease H: the enzymes in eukaryotes. FEBS J. 276, 1494–1505 (2009).
Cerritelli, S. M. et al. Failure to produce mitochondrial DNA results in embryonic lethality in Rnaseh1 null mice. Mol. Cell 11, 807–815 (2003).
Wahba, L., Amon, J. D., Koshland, D. & Vuica-Ross, M. RNase H and multiple RNA biogenesis factors cooperate to prevent RNA:DNA hybrids from generating genome instability. Mol. Cell 44, 978–988 (2011).
Stirling, P. C. et al. R-loop-mediated genome instability in mRNA cleavage and polyadenylation mutants. Genes Dev. 26, 163–175 (2012).
Skourti-Stathaki, K., Kamieniarz-Gdula, K. & Proudfoot, N. J. R-loops induce repressive chromatin marks over mammalian gene terminators. Nature 516, 436–439 (2014). This paper shows a connection between R loops and chromatin repressive marks in the terminator region of human genes. R loops form at these regions and prime antisense transcription, generating dsRNA that seems to be processed by the RNAi machinery. This event triggers H3K9me2 deposition and heterochromatin formation that facilitates RNA Pol II pausing prior to transcription termination.
Boque-Sastre, R. et al. Head-to-head antisense transcription and R-loop formation promotes transcriptional activation. Proc. Natl Acad. Sci. USA 112, 5785–5790 (2015).
Reijns, M. A. et al. Enzymatic removal of ribonucleotides from DNA is essential for mammalian genome integrity and development. Cell 149, 1008–1022 (2012).
Hong, X., Cadwell, G. W. & Kogoma, T. Escherichia coli RecG and RecA proteins in R-loop formation. EMBO J. 14, 2385–2392 (1995).
Harinarayanan, R. & Gowrishankar, J. Host factor titration by chromosomal R-loops as a mechanism for runaway plasmid replication in transcription termination-defective mutants of Escherichia coli. J. Mol. Biol. 332, 31–46 (2003).
Boule, J. B. & Zakian, V. A. The yeast Pif1p DNA helicase preferentially unwinds RNA DNA substrates. Nucleic Acids Res. 35, 5809–5818 (2007).
Chakraborty, P. & Grosse, F. Human DHX9 helicase preferentially unwinds RNA-containing displacement loops (R-loops) and G-quadruplexes. DNA Repair (Amst.) 10, 654–665 (2011).
Kim, H. D., Choe, J. & Seo, Y. S. The sen1+ gene of Schizosaccharomyces pombe, a homologue of budding yeast SEN1, encodes an RNA and DNA helicase. Biochemistry 38, 14697–14710 (1999).
Mischo, H. E. et al. Yeast Sen1 helicase protects the genome from transcription-associated instability. Mol. Cell 41, 21–32 (2011).
Becherel, O. J. et al. Senataxin plays an essential role with DNA damage response proteins in meiotic recombination and gene silencing. PLoS Genet. 9, e1003435 (2013).
De, I. et al. The RNA helicase Aquarius exhibits structural adaptations mediating its recruitment to spliceosomes. Nat. Struct. Mol. Biol. 22, 138–144 (2015).
Sollier, J. et al. Transcription-coupled nucleotide excision repair factors promote R-loop-induced genome instability. Mol. Cell 56, 777–785 (2014). This paper provides a mechanism by which R loops accumulated after depletion of human RNA-processing factors such as AQR may be processed into DSBs by the NER nucleases XPG or XPF. Interestingly, this phenomenon seems to be specific to the transcription-coupled NER sub-pathway.
Paulsen, R. D. et al. A genome-wide siRNA screen reveals diverse cellular processes and pathways that mediate genome stability. Mol. Cell 35, 228–239 (2009).
Drolet, M. et al. Overexpression of RNase H partially complements the growth defect of an Escherichia coli delta topA mutant: R-loop formation is a major problem in the absence of DNA topoisomerase I. Proc. Natl Acad. Sci. USA 92, 3526–3530 (1995).
El Hage, A., French, S. L., Beyer, A. L. & Tollervey, D. Loss of Topoisomerase I leads to R-loop-mediated transcriptional blocks during ribosomal RNA synthesis. Genes Dev. 24, 1546–1558 (2010). This work shows that R loops accumulate in the rDNA of yeast Top1 and Top2 mutants and lead to RNA Pol I transcription impairment, truncated rRNA transcripts and reduced rRNA synthesis. In the absence of TOPs, RNase H activity aids in restoring RNA Pol I transcription efficiency.
Stuckey, R., Garcia-Rodriguez, N., Aguilera, A. & Wellinger, R. E. Role for RNA:DNA hybrids in origin-independent replication priming in a eukaryotic system. Proc. Natl Acad. Sci. USA 112, 5779–5784 (2015).
Tuduri, S. et al. Topoisomerase I suppresses genomic instability by preventing interference between replication and transcription. Nat. Cell Biol. 11, 1315–1324 (2009). This paper shows that TOP1-deficient cells accumulate DNA breaks at transcribed genes all over the genome and have slower replication-fork progression owing to the formation of R loops, suggesting a role for TOP1 in avoiding conflicts between replication and transcription.
Yang, Y. et al. Arginine methylation facilitates the recruitment of TOP3B to chromatin to prevent R loop accumulation. Mol. Cell 53, 484–497 (2014). This work shows that human TOP3B reduces both negative supercoiling and R-loop formation by binding to highly transcribed genes through TDRD3, which recognizes histone and RNA Pol II carboxy-terminal domain methylation. Interestingly, TDRD3-deficient cells accumulate R loops at the MYC locus, and Tdrd3 -null mice show increased Myc–Igh translocations, which are common in Burkitt lymphoma.
Wilson-Sali, T. & Hsieh, T. S. Preferential cleavage of plasmid-based R-loops and D-loops by Drosophila topoisomerase IIIβ. Proc. Natl Acad. Sci. USA 99, 7974–7979 (2002).
Li, M., Pokharel, S., Wang, J. T., Xu, X. & Liu, Y. RECQ5-dependent SUMOylation of DNA topoisomerase I prevents transcription-associated genome instability. Nat. Commun. 6, 6720 (2015).
Castellano-Pozo, M., Garcia-Muse, T. & Aguilera, A. R-loops cause replication impairment and genome instability during meiosis. EMBO Rep. 13, 923–929 (2012).
Dominguez-Sanchez, M. S., Barroso, S., Gomez-Gonzalez, B., Luna, R. & Aguilera, A. Genome instability and transcription elongation impairment in human cells depleted of THO/TREX. PLoS Genet. 7, e1002386 (2011).
Li, X., Niu, T. & Manley, J. L. The RNA binding protein RNPS1 alleviates ASF/SF2 depletion-induced genomic instability. RNA 13, 2108–2115 (2007).
Jimeno, S., Luna, R., Garcia-Rubio, M. & Aguilera, A. Tho1, a novel hnRNP, and Sub2 provide alternative pathways for mRNP biogenesis in yeast THO mutants. Mol. Cell. Biol. 26, 4387–4398 (2006).
Wan, Y. et al. Splicing function of mitotic regulators links R-loop-mediated DNA damage to tumor cell killing. J. Cell Biol. 209, 235–246 (2015).
Salvi, J. S. et al. Roles for Pbp1 and caloric restriction in genome and lifespan maintenance via suppression of RNA-DNA hybrids. Dev. Cell 30, 177–191 (2014).
Gavalda, S., Gallardo, M., Luna, R. & Aguilera, A. R-loop mediated transcription-associated recombination in trf4Δ mutants reveals new links between RNA surveillance and genome integrity. PLoS ONE 8, e65541 (2013).
Pefanis, E. et al. RNA exosome-regulated long non-coding RNA transcription controls super-enhancer activity. Cell 161, 774–789 (2015).
Sun, Q., Csorba, T., Skourti-Stathaki, K., Proudfoot, N. J. & Dean, C. R-loop stabilization represses antisense transcription at the Arabidopsis FLC locus. Science 340, 619–621 (2013).
Proudfoot, N. J. Ending the message: poly(A) signals then and now. Genes Dev. 25, 1770–1782 (2011).
Kawauchi, J., Mischo, H., Braglia, P., Rondon, A. & Proudfoot, N. J. Budding yeast RNA polymerases I and II employ parallel mechanisms of transcriptional termination. Genes Dev. 22, 1082–1092 (2008).
Grzechnik, P., Gdula, M. R. & Proudfoot, N. J. Pcf11 orchestrates transcription termination pathways in yeast. Genes Dev. 29, 849–861 (2015).
Porrua, O. & Libri, D. A bacterial-like mechanism for transcription termination by the Sen1p helicase in budding yeast. Nat. Struct. Mol. Biol. 20, 884–891 (2013).
Yuce, O. & West, S. C. Senataxin, defective in the neurodegenerative disorder ataxia with oculomotor apraxia 2, lies at the interface of transcription and the DNA damage response. Mol. Cell. Biol. 33, 406–417 (2013).
Morales, J. C. et al. Kub5-Hera, the human Rtt103 homolog, plays dual functional roles in transcription termination and DNA repair. Nucleic Acids Res. 42, 4996–5006 (2014).
Huang, H. S. et al. Topoisomerase inhibitors unsilence the dormant allele of Ube3a in neurons. Nature 481, 185–189 (2012).
Powell, W. T. et al. R-loop formation at Snord116 mediates topotecan inhibition of Ube3a-antisense and allele-specific chromatin decondensation. Proc. Natl Acad. Sci. USA 110, 13938–13943 (2013).
Lindahl, T. Instability and decay of the primary structure of DNA. Nature 362, 709–715 (1993).
Polak, P. & Arndt, P. F. Transcription induces strand-specific mutations at the 5′ end of human genes. Genome Res. 18, 1216–1223 (2008).
Chaudhuri, J. & Alt, F. W. Class-switch recombination: interplay of transcription, DNA deamination and DNA repair. Nat. Rev. Immunol. 4, 541–552 (2004).
Meng, F. L. et al. Convergent transcription at intragenic super-enhancers targets AID-initiated genomic instability. Cell 159, 1538–1548 (2014).
Qian, J. et al. B cell super-enhancers and regulatory clusters recruit AID tumorigenic activity. Cell 159, 1524–1537 (2014).
Gómez-González, B. & Aguilera, A. Activation-induced cytidine deaminase action is strongly stimulated by mutations of the THO complex. Proc. Natl Acad. Sci. USA 104, 8409–8414 (2007).
Kogoma, T. Stable DNA replication: interplay between DNA replication, homologous recombination, and transcription. Microbiol. Mol. Biol. Rev. 61, 212–238 (1997).
Ponder, R. G., Fonville, N. C. & Rosenberg, S. M. A switch from high-fidelity to error-prone DNA double-strand break repair underlies stress-induced mutation. Mol. Cell 19, 791–804 (2005).
Wimberly, H. et al. R-loops and nicks initiate DNA breakage and genome instability in non-growing Escherichia coli. Nat. Commun. 4, 2115 (2013).
Deem, A. et al. Break-induced replication is highly inaccurate. PLoS Biol. 9, e1000594 (2011).
Aguilera, A. & García-Muse, T. Causes of genome instability. Annu. Rev. Genet. 47, 1–32 (2013).
Boubakri, H., de Septenville, A. L., Viguera, E. & Michel, B. The helicases DinG, Rep and UvrD cooperate to promote replication across transcription units in vivo. EMBO J. 29, 145–157 (2010).
Gan, W. et al. R-loop-mediated genomic instability is caused by impairment of replication fork progression. Genes Dev. 25, 2041–2056 (2011).
Wellinger, R. E., Prado, F. & Aguilera, A. Replication fork progression is impaired by transcription in hyperrecombinant yeast cells lacking a functional THO complex. Mol. Cell. Biol. 26, 3327–3334 (2006).
Gómez-González, B. et al. Genome-wide function of THO/TREX in active genes prevents R-loop-dependent replication obstacles. EMBO J. 30, 3106–3119 (2011).
Santos-Pereira, J. M. et al. The Npl3 hnRNP prevents R-loop-mediated transcription-replication conflicts and genome instability. Genes Dev. 27, 2445–2458 (2013).
Alzu, A. et al. Senataxin associates with replication forks to protect fork integrity across RNA-polymerase-II-transcribed genes. Cell 151, 835–846 (2012). This article provides genome-wide evidence that Sen1 (the yeast orthologue of the mammalian RNA–DNA helicase senataxin) accumulates with replication forks at transcribed genes, suggesting that Sen1 protects forks from the formation of recombinogenic damage that can activate the DNA damage checkpoint.
Helmrich, A., Ballarino, M. & Tora, L. Collisions between replication and transcription complexes cause common fragile site instability at the longest human genes. Mol. Cell 44, 966–977 (2011). In this work, the authors show that the longest human genes take more than a cell cycle to be transcribed and that collision between transcription and replication machineries is inevitable. This leads to hotspots of DNA breaks called common fragile sites, where R loops form and are responsible for the transcription–replication conflicts that generate genome instability.
Róndon, A. G., Jimeno, S., García-Rubio, M. & Aguilera, A. Molecular evidence that the eukaryotic THO/TREX complex is required for efficient transcription elongation. J. Biol. Chem. 278, 39037–39043 (2003).
Dutta, D., Shatalin, K., Epshtein, V., Gottesman, M. E. & Nudler, E. Linking RNA polymerase backtracking to genome instability in E. coli. Cell 146, 533–543 (2011).
Tresini, M. et al. The core spliceosome as target and effector of non-canonical ATM signalling. Nature 523, 53–58 (2015).
Bhatia, V. et al. BRCA2 prevents R-loop accumulation and associates with TREX-2 mRNA export factor PCID2. Nature 511, 362–365 (2014). This paper shows that depletion of BRCA2 leads to genome instability and accumulation of R loops, as detected by the S9.6 antibody, which specifically recognizes these structures, and by an RNase H1 hybrid-binding domain fused to GFP. The manuscript proposes that R loops are a major source of spontaneous replication stress and that BRCA2 and Fanconi anaemia proteins contribute to the elimination of R loops that block replication-fork progression.
Schlacher, K., Wu, H. & Jasin, M. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell 22, 106–116 (2012).
Britton, S. et al. DNA damage triggers SAF-A and RNA biogenesis factors exclusion from chromatin coupled to R-loops removal. Nucleic Acids Res. 42, 9047–9062 (2014).
Luke, B. et al. The Rat1p 5′ to 3′ exonuclease degrades telomeric repeat-containing RNA and promotes telomere elongation in Saccharomyces cerevisiae. Mol. Cell 32, 465–477 (2008).
Balk, B. et al. Telomeric RNA-DNA hybrids affect telomere-length dynamics and senescence. Nat. Struct. Mol. Biol. 20, 1199–1205 (2013).
Pfeiffer, V., Crittin, J., Grolimund, L. & Lingner, J. The THO complex component Thp2 counteracts telomeric R-loops and telomere shortening. EMBO J. 32, 2861–2871 (2013).
Yu, T. Y., Kao, Y. W. & Lin, J. J. Telomeric transcripts stimulate telomere recombination to suppress senescence in cells lacking telomerase. Proc. Natl Acad. Sci. USA 111, 3377–3382 (2014).
Arora, R. et al. RNaseH1 regulates TERRA- telomeric DNA hybrids and telomere maintenance in ALT tumour cells. Nat. Commun. 5, 5220 (2014).
Nakama, M., Kawakami, K., Kajitani, T., Urano, T. & Murakami, Y. DNA–RNA hybrid formation mediates RNAi-directed heterochromatin formation. Genes Cells 17, 218–233 (2012).
Hsu, J. Y. et al. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell 102, 279–291 (2000).
Ivaldi, M. S., Karam, C. S. & Corces, V. G. Phosphorylation of histone H3 at Ser10 facilitates RNA polymerase II release from promoter-proximal pausing in Drosophila. Genes Dev. 21, 2818–2831 (2007).
Zippo, A., De Robertis, A., Serafini, R. & Oliviero, S. PIM1-dependent phosphorylation of histone H3 at serine 10 is required for MYC-dependent transcriptional activation and oncogenic transformation. Nat. Cell Biol. 9, 932–944 (2007).
El Achkar, E., Gerbault-Seureau, M., Muleris, M., Dutrillaux, B. & Debatisse, M. Premature condensation induces breaks at the interface of early and late replicating chromosome bands bearing common fragile sites. Proc. Natl Acad. Sci. USA 102, 18069–18074 (2005).
Groh, M., Lufino, M. M., Wade-Martins, R. & Gromak, N. R-loops associated with triplet repeat expansions promote gene silencing in Friedreich ataxia and fragile X syndrome. PLoS Genet. 10, e1004318 (2014). These authors used cells from people with FRDA or FXS, diseases that are characterized by the expansion of trinucleotides in the FXN and FMR1 genes, respectively. These expansions constitute rare fragile sites, and the article shows that R loops form at these expanded repeats, leading to accumulation of the repressive mark H3K9me2 and consequent gene silencing, which causes the disease.
Herrera-Moyano, E., Mergui, X., Garcia-Rubio, M. L., Barroso, S. & Aguilera, A. The yeast and human FACT chromatin-reorganizing complexes solve R-loop-mediated transcription-replication conflicts. Genes Dev. 28, 735–748 (2014).
Hatchi, E. et al. BRCA1 recruitment to transcriptional pause sites is required for R-loop-driven DNA damage repair. Mol. Cell 57, 636–647 (2015). This work suggests a cooperation between the human DNA repair factor BRCA1 and the RNA–DNA helicase SETX at transcription-termination regions to prevent R-loop formation, as depletion of either factor leads to increased levels of R loops. Interestingly, BRCA1 and SETX physically interact, and SETX binding to termination regions is BRCA1-dependent. BRCA1 binds genome-wide to termination regions of R-loop-accumulating genes, where BRCA1-deficient tumours show increased insertions and deletions.
Lin, Y., Dent, S. Y., Wilson, J. H., Wells, R. D. & Napierala, M. R loops stimulate genetic instability of CTG. CAG repeats. Proc. Natl Acad. Sci. USA 107, 692–697 (2010).
Reddy, K. et al. Determinants of R-loop formation at convergent bidirectionally transcribed trinucleotide repeats. Nucleic Acids Res. 39, 1749–1762 (2011).
Grabczyk, E., Mancuso, M. & Sammarco, M. C. A persistent RNA•DNA hybrid formed by transcription of the Friedreich ataxia triplet repeat in live bacteria, and by T7 RNAP in vitro. Nucleic Acids Res. 35, 5351–5359 (2007).
Loomis, E. W., Sanz, L. A., Chedin, F. & Hagerman, P. J. Transcription-associated R-loop formation across the human FMR1 CGG-repeat region. PLoS Genet. 10, e1004294 (2014).
Colak, D. et al. Promoter-bound trinucleotide repeat mRNA drives epigenetic silencing in fragile X syndrome. Science 343, 1002–1005 (2014).
Haeusler, A. R. et al. C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature 507, 195–200 (2014).
Moreira, M. C. et al. Senataxin, the ortholog of a yeast RNA helicase, is mutant in ataxia- ocular apraxia 2. Nat. Genet. 36, 225–227 (2004).
Chen, Y. Z. et al. DNA/RNA helicase gene mutations in a form of juvenile amyotrophic lateral sclerosis (ALS4). Am. J. Hum. Genet. 74, 1128–1135 (2004).
Vantaggiato, C. et al. Senataxin modulates neurite growth through fibroblast growth factor 8 signalling. Brain 134, 1808–1828 (2011).
Yeo, A. J. et al. R-loops in proliferating cells but not in the brain: implications for AOA2 and other autosomal recessive ataxias. PLoS ONE 9, e90219 (2014).
Negrini, S., Gorgoulis, V. G. & Halazonetis, T. D. Genomic instability — an evolving hallmark of cancer. Nat. Rev. Mol. Cell Biol. 11, 220–228 (2010).
Gaillard, H., Garcia-Muse, T. & Aguilera, A. Replication stress and cancer. Nat. Rev. Cancer 15, 276–289 (2015).
Hill, S. J. et al. Systematic screening reveals a role for BRCA1 in the response to transcription-associated DNA damage. Genes Dev. 28, 1957–1975 (2014).
Ramiro, A. R. et al. AID is required for c-myc/IgH chromosome translocations in vivo. Cell 118, 431–438 (2004).
Ruiz, J. F., Gómez-González, B. & Aguilera, A. AID induces double-strand breaks at immunoglobulin switch regions and c-MYC causing chromosomal translocations in yeast THO mutants. PLoS Genet. 7, e1002009 (2011).
Chernikova, S. B. et al. Deficiency in mammalian histone H2B ubiquitin ligase Bre1 (Rnf20/Rnf40) leads to replication stress and chromosomal instability. Cancer Res. 72, 2111–2119 (2012).
Fregoso, O. I., Das, S., Akerman, M. & Krainer, A. R. Splicing-factor oncoprotein SRSF1 stabilizes p53 via RPL5 and induces cellular senescence. Mol. Cell 50, 56–66 (2013).
Jackson, B. R., Noerenberg, M. & Whitehouse, A. A novel mechanism inducing genome instability in Kaposi's sarcoma-associated herpesvirus infected cells. PLoS Pathog. 10, e1004098 (2014).
Huang, F. T. et al. Sequence dependence of chromosomal R-loops at the immunoglobulin heavy-chain Smu class switch region. Mol. Cell. Biol. 27, 5921–5932 (2007).
Huang, F. T., Yu, K., Hsieh, C. L. & Lieber, M. R. Downstream boundary of chromosomal R-loops at murine switch regions: implications for the mechanism of class switch recombination. Proc. Natl Acad. Sci. USA 103, 5030–5035 (2006).
Kao, Y. P. et al. Detection and characterization of R-loops at the murine immunoglobulin Sα region. Mol. Immunol. 54, 208–216 (2013).
Yu, K., Chedin, F., Hsieh, C. L., Wilson, T. E. & Lieber, M. R. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat. Immunol. 4, 442–451 (2003).
Reaban, M. E. & Griffin, J. A. Induction of RNA-stabilized DNA conformers by transcription of an immunoglobulin switch region. Nature 348, 342–344 (1990).
Muramatsu, M. et al. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell 102, 553–563 (2000).
Revy, P. et al. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2). Cell 102, 565–575 (2000).
Chaudhuri, J. et al. Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature 422, 726–730 (2003).
Zheng, S. et al. Non-coding RNA generated following lariat debranching mediates targeting of AID to DNA. Cell 161, 762–773 (2015).
Pefanis, E. et al. Noncoding RNA transcription targets AID to divergently transcribed loci in B cells. Nature 514, 389–393 (2014).
Zhang, Z. Z. et al. The strength of an Ig switch region is determined by its ability to drive R loop formation and its number of WGCW sites. Cell Rep. 8, 557–569 (2014).
Boguslawski, S. J. et al. Characterization of monoclonal antibody to DNA•RNA and its application to immunodetection of hybrids. J. Immunol. Methods 89, 123–130 (1986).
Zhang, Z. Z., Pannunzio, N. R., Hsieh, C. L., Yu, K. & Lieber, M. R. Complexities due to single-stranded RNA during antibody detection of genomic rna:dna hybrids. BMC Res. Notes 8, 127 (2015).
Jenjaroenpun, P., Wongsurawat, T., Yenamandra, S. P. & Kuznetsov, V. A. QmRLFS-finder: a model, web server and stand-alone tool for prediction and analysis of R-loop forming sequences. Nucleic Acids Res. 43, W527–W534 (2015).
Leela, J. K., Syeda, A. H., Anupama, K. & Gowrishankar, J. Rho-dependent transcription termination is essential to prevent excessive genome-wide R-loops in Escherichia coli. Proc. Natl Acad. Sci. USA 110, 258–263 (2013).
Wahba, L., Gore, S. K. & Koshland, D. The homologous recombination machinery modulates the formation of RNA•DNA hybrids and associated chromosome instability. eLife 2, e00505 (2013).
Acknowledgements
The authors thank T. García-Muse for critical reading of the manuscript and D. Haun for style supervision. Research in A.A.'s laboratory is funded by grants from the Spanish Ministry of Economy and Competitiveness, Junta de Andalucía, European Union (FEDER), Worldwide Cancer Research and PharmaMar. The authors apologize to those whose work could not be cited owing to space limitations.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Glossary
- G-quartets
-
Planar structures formed by four intra-strand Gs paired with each other.
- Genome instability
-
A cellular pathological condition by which cells undergo mutations, recombination, chromosome rearrangements or chromosome loss at frequencies substantially higher than wild-type levels.
- Holliday junctions
-
Double-stranded, cruciform structures formed as intermediates of recombinational repair in which both recombining DNA molecules are covalently linked.
- Recombinational repair
-
A double-strand break repair that is active during the S–G2 phases of the cell cycle and that uses information from a homologous sequence, normally the sister chromatid, to copy DNA.
- Okazaki fragment
-
A discrete fragment created by synthesis of the DNA lagging strand during replication. Okazaki fragments are primed by a short RNA in the form of an RNA–DNA hybrid.
- Negative supercoiling
-
Under-winding of a DNA strand, typically occurring behind an elongating RNA polymerase.
- Topoisomerase
-
(TOP). An enzyme family that can remove (or create) supercoiling in duplex DNA by making transitory breaks in one strand (type 1 TOPs) or both strands (type 2 TOPs) of the DNA backbone.
- D loops
-
(Displacement loops). DNA structures consisting of a main double-stranded DNA that has been separated by a third DNA strand complementary to one of the main strands and that by pairing with it displaces the other main strand.
- Bisulfite mutagenesis
-
A method to mutagenize DNA with sodium bisulfite, which acts exclusively over single-stranded DNA and, therefore, serves to determine whether a particular DNA sequence is present in vivo in a single-stranded form (as is the case for the strand displaced by the RNA in the R loop) or in a double-stranded form.
- DNA damage response
-
(DDR). A general cellular response consisting of DNA damage sensing, activation of different checkpoints, the action of the appropriate DNA repair pathway and the arrest of the cell cycle.
- Multicopy suppressors
-
Genes that are able to suppress the phenotype conferred by specific mutations when they are present in high-copy-number plasmids.
- CpG islands
-
CpG-rich regions that are usually unmethylated and localized at the 5′ end of genes, where they function as promoter elements.
- GC skew
-
Asymmetry in the distribution of Gs and Cs between DNA strands, with an overrepresentation of Gs in the non-transcribed DNA strand.
- Heterochromatin
-
A chromosomal region with highly compacted chromatin that is more refractory to the action of enzymes, has a general repressive action on gene transcription and replicates late in the cell cycle.
- Homeodomain
-
A DNA-binding domain that is characteristic of homeobox proteins involved in transcription regulation. It consists of a 60-amino-acid helix–turn–helix structure with three α-helices connected by loop regions.
- RNA interference
-
(RNAi). A mechanism of gene silencing that relies on short non-coding RNAs that have the ability to repress chromatin with the help of additional ancillary factors.
- Replication stress
-
Any condition in which replication progression slows down and/or stalls, commonly leading to genome instability.
- Break-induced replication
-
(BIR). A mechanism of recombinational repair in which a one-ended double-strand break invades a homologous DNA sequence that is used as template for DNA synthesis to complete repair.
- DNA combing
-
A technique used to produce stretched DNA fibres for multiple applications, including the study of DNA replication by immune detection of modified nucleotides.
- Common fragile site
-
A specific chromosome region that has gaps or constrictions that are visible under the microscope and that tend to break on exposure to replication stress.
- Backtracked RNA polymerase
-
An intermediate state in which an arrested RNA polymerase moves back to allow cleavage of the last ribonucleotide incorporated into the nascent RNA, thus allowing transcription resumption.
- Nucleotide excision repair
-
(NER). A conserved DNA repair pathway that recognizes adducts and repairs them by excision of a short oligonucleotide containing the damage.
- Fanconi anaemia pathway
-
A DNA repair pathway that works on replication forks stalled at interstrand crosslinks and other lesions that block replication.
Rights and permissions
About this article
Cite this article
Santos-Pereira, J., Aguilera, A. R loops: new modulators of genome dynamics and function. Nat Rev Genet 16, 583–597 (2015). https://doi.org/10.1038/nrg3961
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrg3961
This article is cited by
-
Dual roles of R-loops in the formation and processing of programmed DNA double-strand breaks during meiosis
Cell & Bioscience (2023)
-
R-loop-derived cytoplasmic RNA–DNA hybrids activate an immune response
Nature (2023)
-
APOBEC3B coordinates R-loop to promote replication stress and sensitize cancer cells to ATR/Chk1 inhibitors
Cell Death & Disease (2023)
-
Splicing factor SRSF1 deficiency in the liver triggers NASH-like pathology and cell death
Nature Communications (2023)
-
R-loop-dependent promoter-proximal termination ensures genome stability
Nature (2023)