Key Points
-
Technological advances have vastly expanded the amount of omic data currently available. Historically, each type of data was analysed separately, although approaches to integrate omic data sets to predict complex phenotypic traits are now emerging.
-
Such systems genomics approaches to combine multiple data types provide a more comprehensive understanding of complex genotype–phenotype associations than analysis of one data set.
-
Data from multiple sources that point to the association of the same gene or pathway are less likely to result in false positives.
-
There are various strengths and weaknesses of the available strategies. The approach used needs to be selected according to specific types of data, different types of scientific questions or different types of underlying genomic models.
Abstract
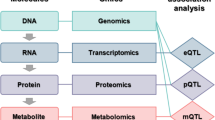
Recent technological advances have expanded the breadth of available omic data, from whole-genome sequencing data, to extensive transcriptomic, methylomic and metabolomic data. A key goal of analyses of these data is the identification of effective models that predict phenotypic traits and outcomes, elucidating important biomarkers and generating important insights into the genetic underpinnings of the heritability of complex traits. There is still a need for powerful and advanced analysis strategies to fully harness the utility of these comprehensive high-throughput data, identifying true associations and reducing the number of false associations. In this Review, we explore the emerging approaches for data integration — including meta-dimensional and multi-staged analyses — which aim to deepen our understanding of the role of genetics and genomics in complex outcomes. With the use and further development of these approaches, an improved understanding of the relationship between genomic variation and human phenotypes may be revealed.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Metzker, M. L. Sequencing technologies — the next generation. Nature Rev. Genet. 11, 31–46 (2010).
Ozsolak, F. & Milos, P. M. RNA sequencing: advances, challenges and opportunities. Nature Rev. Genet. 12, 87–98 (2011).
Wang, Z., Gerstein, M. & Snyder, M. RNA-Seq: a revolutionary tool for transcriptomics. Nature Rev. Genet. 10, 57–63 (2009).
Laird, P. W. Principles and challenges of genome-wide DNA methylation analysis. Nature Rev. Genet. 11, 191–203 (2010). This is a comprehensive review of DNA methylation data analysis.
Park, P. J. ChIP–seq: advantages and challenges of a maturing technology. Nature Rev. Genet. 10, 669–680 (2009).
Altelaar, A. F. M., Munoz, J. & Heck, A. J. R. Next-generation proteomics: towards an integrative view of proteome dynamics. Nature Rev. Genet. 14, 35–48 (2013).
Shulaev, V. Metabolomics technology and bioinformatics. Brief. Bioinform. 7, 128–139 (2006).
Shapiro, E., Biezuner, T. & Linnarsson, S. Single-cell sequencing-based technologies will revolutionize whole-organism science. Nature Rev. Genet. 14, 618–630 (2013).
Almasy, L. & Blangero, J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am. J. Hum. Genet. 62, 1198–1211 (1998).
Horvath, S., Xu, X. & Laird, N. M. The family based association test method: strategies for studying general genotype—phenotype associations. Eur. J. Hum. Genet. 9, 301–306 (2001).
Devlin, B., Roeder, K. & Bacanu, S. A. Unbiased methods for population-based association studies. Genet. Epidemiol. 21, 273–284 (2001).
Reif, D. M., White, B. C. & Moore, J. H. Integrated analysis of genetic, genomic and proteomic data. Expert Rev. Proteomics 1, 67–75 (2004).
Hamid, J. S. et al. Data integration in genetics and genomics: methods and challenges. Hum. Genomics Proteomics 2009, 869093 (2009).
Sieberts, S. K. & Schadt, E. E. Moving toward a system genetics view of disease. Mamm. Genome 18, 389–401 (2007).
Hawkins, R. D., Hon, G. C. & Ren, B. Next-generation genomics: an integrative approach. Nature Rev. Genet. 11, 476–486 (2010).
Holzinger, E. R. & Ritchie, M. D. Integrating heterogeneous high-throughput data for meta-dimensional pharmacogenomics and disease-related studies. Pharmacogenomics 13, 213–222 (2012).
Holzinger, E. et al. in Evolutionary Computation, Machine Learning and Data Mining in Bioinformatics (eds Giacobini, M., Vanneschi, L. & Bush, W.) 7246, 134–143 (Springer Berlin Heidelberg, 2012).
Holzinger, E. R. et al. ATHENA: a tool for meta-dimensional analysis applied to genotypes and gene expression data to predict HDL cholesterol levels. Pac. Symp. Biocomput. 385–396 (2013).
Stein, L. D. The case for cloud computing in genome informatics. Genome Biol. 11, 207 (2010).
Dorff, K. C. et al. GobyWeb: simplified management and analysis of gene expression and DNA methylation sequencing data. PLoS ONE 8, e69666 (2013).
Reid, J. G. et al. Launching genomics into the cloud: deployment of Mercury, a next generation sequence analysis pipeline. BMC Bioinformatics 15, 30 (2014).
Heath, A. P. et al. Bionimbus: a cloud for managing, analyzing and sharing large genomics datasets. J. Am. Med. Inform. Assoc. 21, 969–975 (2014).
Turner, S. et al. Quality control procedures for genome-wide association studies. Curr. Protoc. Hum. Genet. 68, 1.19.1–1.19.18 (2011).
Zuvich, R. L. et al. Pitfalls of merging GWAS data: lessons learned in the eMERGE network and quality control procedures to maintain high data quality. Genet. Epidemiol. 35, 887–898 (2011). This paper provides detailed lessons learned about quality control processes in high-throughput genotype data and guides readers toward best practices when cleaning and merging genotype data.
Laurie, C. C. et al. Quality control and quality assurance in genotypic data for genome-wide association studies. Genet. Epidemiol. 34, 591–602 (2010).
McKenna, A. et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
Marguerat, S. & Bähler, J. RNA-seq: from technology to biology. Cell. Mol. Life Sci. 67, 569–579 (2010).
Hirst, M. & Marra, M. A. Next generation sequencing based approaches to epigenomics. Briefings Funct. Genom. 9, 455–465 (2010).
Johnstone, I. M. & Titterington, D. M. Statistical challenges of high-dimensional data. Phil. Trans. R. Soc. A. 367, 4237–4253 (2009).
Hastie, T., Tibshirani, R. & Friedman, J. The Elements of Statistical Learning: Data Mining, Inference, and Prediction (Springer-Verlag, 2001).
Bush, W. S., Dudek, S. M. & Ritchie, M. D. Biofilter: a knowledge-integration system for the multi-locus analysis of genome-wide association studies. Pac. Symp. Biocomput. 368–379 (2009).
Greene, C. S., Penrod, N. M., Kiralis, J. & Moore, J. H. Spatially uniform ReliefF (SURF) for computationally-efficient filtering of gene–gene interactions. BioData Min. 2, 5 (2009).
Moore, J. H. & White, B. C. in Evolutionary Computation, Machine Learning and Data Mining in Bioinformatics (eds Marchiori, E., Moore, J. H. & Rajapakse, J. C.) 166–175 (Springer Berlin Heidelberg, 2007).
Zou, H., Hastie, T. & Tibshirani, R. Sparse principal component analysis. J. Comput. Graph. Stat. 15, 265–286 (2006).
Holland, J. H. Genetic algorithms. Sci. Am. 267, 66–72 (1992).
Vilhjálmsson, B. J. & Nordborg, M. The nature of confounding in genome-wide association studies. Nature Rev. Genet. 14, 1–2 (2013).
Zhou, X. & Stephens, M. Efficient multivariate linear mixed model algorithms for genome-wide association studies. Nature Methods 11, 407–409 (2014).
Price, A. L. et al. Principal components analysis corrects for stratification in genome-wide association studies. Nature Genet. 38, 904–909 (2006).
Leek, J. T. & Storey, J. D. Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet. 3, e161 (2007).
Hartford, C. M. et al. Population-specific genetic variants important in susceptibility to cytarabine arabinoside cytotoxicity. Blood 113, 2145–2153 (2009).
Huang, R. S. et al. A genome-wide approach to identify genetic variants that contribute to etoposide-induced cytotoxicity. Proc. Natl Acad. Sci. USA 104, 9758–9763 (2007). This is one of the first papers to present an integrative analysis to identify DNA variants and gene expressions associated with chemotherapeutic drug-induced cytotoxicity.
Huang, R. S., Duan, S., Kistner, E. O., Hartford, C. M. & Dolan, M. E. Genetic variants associated with carboplatin-induced cytotoxicity in cell lines derived from Africans. Mol. Cancer Ther. 7, 3038–3046 (2008).
Schadt, E. E. et al. An integrative genomics approach to infer causal associations between gene expression and disease. Nature Genet. 37, 710–717 (2005). This study used an integrative approach to use DNA variation and gene expression data to identify drivers of complex traits.
Liu, Y. et al. Epigenome-wide association data implicate DNA methylation as an intermediary of genetic risk in rheumatoid arthritis. Nature Biotech. 31, 142–147 (2013).
Khan, Z. et al. Quantitative measurement of allele-specific protein expression in a diploid yeast hybrid by LC-MS. Mol. Syst. Biol. 8, 602 (2012).
Wei, X. & Wang, X. A computational workflow to identify allele-specific expression and epigenetic modification in maize. Genomics Proteomics Bioinformatics 11, 247–252 (2013).
Lappalainen, T. et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature 501, 506–511 (2013). This paper reports the sequencing and analysis of mRNA and microRNA of hundreds of multi-ethnic individuals from the 1000 Genome Project.
Maynard, N. D., Chen, J., Stuart, R. K., Fan, J.-B. & Ren, B. Genome-wide mapping of allele-specific protein–DNA interactions in human cells. Nature Methods 5, 307–309 (2008).
Kasowski, M. et al. Extensive variation in chromatin states across humans. Science 342, 750–752 (2013).
McVicker, G. et al. Identification of genetic variants that affect histone modifications in human cells. Science 342, 747–749 (2013).
Encode Project Consortium. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science 306, 636–640 (2004).
Kanehisa, M. & Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 (2000).
Kim, D., Shin, H., Song, Y. S. & Kim, J. H. Synergistic effect of different levels of genomic data for cancer clinical outcome prediction. J. Biomed. Inform. 45, 1191–1198 (2012). This study shows a graph-based approach for predicting cancer clinical outcome by integrating multi-omics data as a transformation-based integration.
Fridley, B. L., Lund, S., Jenkins, G. D. & Wang, L. A. Bayesian integrative genomic model for pathway analysis of complex traits. Genet. Epidemiol. 36, 352–359 (2012).
Mankoo, P. K., Shen, R., Schultz, N., Levine, D. A. & Sander, C. Time to recurrence and survival in serous ovarian tumors predicted from integrated genomic profiles. PLoS ONE 6, e24709 (2011).
Holzinger, E. R., Dudek, S. M., Frase, A. T., Pendergrass, S. A. & Ritchie, M. D. ATHENA: the analysis tool for heritable and environmental network associations. Bioinformatics 30, 698–705 (2014). ATHENA is a tool for meta-dimensional integration of multi-omics data. This paper describes the software and its application for these types of analyses.
Kim, D., Li, R., Dudek, S. M. & Ritchie, M. D. ATHENA: Identifying interactions between different levels of genomic data associated with cancer clinical outcomes using grammatical evolution neural network. BioData Min. 6, 23 (2013).
Clarke, R. et al. The properties of high-dimensional data spaces: implications for exploring gene and protein expression data. Nature Rev. Cancer 8, 37–49 (2008). This review addresses the properties of high-dimensional data spaces and the challenges for data analysis and interpretation.
Lanckriet, G. R. G., De Bie, T., Cristianini, N., Jordan, M. I. & Noble, W. S. A statistical framework for genomic data fusion. Bioinformatics 20, 2626–2635 (2004). This is the first study to propose a kernel-based integration as a transformation-based integration.
Borgwardt, K. M. et al. Protein function prediction via graph kernels. Bioinformatics 21, i47–i56 (2005).
Tsuda, K., Shin, H. & Schölkopf, B. Fast protein classification with multiple networks. Bioinformatics 21, ii59–ii65 (2005).
Shin, H., Lisewski, A. M. & Lichtarge, O. Graph sharpening plus graph integration: a synergy that improves protein functional classification. Bioinformatics 23, 3217–3224 (2007).
Turner, S. D., Dudek, S. M. & Ritchie, M. D. ATHENA: a knowledge-based hybrid backpropagation-grammatical evolution neural network algorithm for discovering epistasis among quantitative trait loci. BioData Min. 3, 5 (2010).
Dra˘ghici, S. & Potter, R. B. Predicting HIV drug resistance with neural networks. Bioinformatics 19, 98–107 (2003).
Shen, H.-B. & Chou, K.-C. Ensemble classifier for protein fold pattern recognition. Bioinformatics 22, 1717–1722 (2006).
Akavia, U. D. et al. An integrated approach to uncover drivers of cancer. Cell 143, 1005–1017 (2010). This paper demonstrated a computational framework that identified drivers of melanoma using chromosomal copy number and gene expression data.
Zhu, J. et al. Stitching together multiple data dimensions reveals interacting metabolomic and transcriptomic networks that modulate cell regulation. PLoS Biol. 10, e1001301 (2012).
Zhu, J. et al. Integrating large-scale functional genomic data to dissect the complexity of yeast regulatory networks. Nature Genet. 40, 854–861 (2008).
Opitz, D. & Maclin, R. Popular ensemble methods: an empirical study. J. Artif. Intell. Res. 11, 169–198 (1999).
Shen, R. et al. Integrative subtype discovery in glioblastoma using iCluster. PLoS ONE 7, e35236 (2012).
Kirk, P., Griffin, J. E., Savage, R. S., Ghahramani, Z. & Wild, D. L. Bayesian correlated clustering to integrate multiple datasets. Bioinformatics 28, 3290–3297 (2012).
Lock, E. F. & Dunson, D. B. Bayesian consensus clustering. Bioinformatics 29, 2610–2616 (2013).
Dupont, W. D. & Plummer, W. D. Power and sample size calculations. A review and computer program. Control Clin. Trials 11, 116–128 (1990).
NCI–NHGRI Working Group on Replication in Association Studies. Replicating genotype–phenotype associations. Nature 447, 655–660 (2007).
Greene, C. S., Penrod, N. M., Williams, S. M. & Moore, J. H. Failure to replicate a genetic association may provide important clues about genetic architecture. PLoS ONE 4, e5639 (2009).
Ciesielski, T. et al. Diverse convergent evidence in the genetic analysis of complex disease: Coordinating omic, informatic, and experimental evidence to better identify and validate risk factors. BioData Min. 7, 10 (2014).
Van Poucke, M., Vanhaesebrouck, A. E., Peelman, L. J. & Van Ham, L. Experimental validation of in silico predicted KCNA1, KCNA2, KCNA6 and KCNQ2 genes for association studies of peripheral nerve hyperexcitability syndrome in Jack Russell Terriers. Neuromuscul. Disord. 22, 558–565 (2012).
Sharaf, R. N. et al. Computational prediction and experimental validation associating FABP-1 and pancreatic adenocarcinoma with diabetes. BMC Gastroenterol. 11, 5 (2011).
Raychaudhuri, S. et al. Identifying relationships among genomic disease regions: predicting genes at pathogenic SNP associations and rare deletions. PLoS Genet. 5, e1000534 (2009).
Crooke, P. S. et al. Estrogens, enzyme variants, and breast cancer: a risk model. Cancer Epidemiol. Biomarkers Prev. 15, 1620–1629 (2006).
Farrar, D. E. & Glauber, R. R. Multicollinearity in regression analysis: the problem revisited. Rev. Econ. Stat. 49, 92 (1967).
Fawcett, T. An introduction to ROC analysis. Pattern Recogn. Lett. 27, 861–874 (2006).
Moore, J.H., Hill, D. P., Sulovari, A & Kidd, L.C. in Genetic Programming Theory and Practice X 87–101 (Springer, 2013).
Jin, Y. & Sendhoff, B. Pareto-based multiobjective machine learn: an overview case studies. IEEE Trans. Syst. Man Cybern. C Appl. Rev. 38, 397–415 (2008).
Kristensen, V. N. & Borresen-Dale, A. L. Molecular epidemiology of breast cancer: genetic variation in steroid hormone metabolism. Mutat. Res. 462, 323–333 (2000).
Mitrunen, K. et al. Glutathione S-transferase M1, M3, P1, and T1 genetic polymorphisms and susceptibility to breast cancer. Cancer Epidemiol. Biomarkers Prev. 10, 229–236 (2001).
Kiyotani, K. et al. A genome-wide association study identifies locus at 10q22 associated with clinical outcomes of adjuvant tamoxifen therapy for breast cancer patients in Japanese. Hum. Mol. Genet. 21, 1665–1672 (2012).
Garcia-Closas, M. et al. Genome-wide association studies identify four ER negative-specific breast cancer risk loci. Nature Genet. 45, 392–398, 398e1–2 (2013).
Michailidou, K. et al. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nature Genet. 45, 353–361, 361e1–2 (2013).
Zheng, W. et al. Common genetic determinants of breast-cancer risk in East Asian women: a collaborative study of 23 637 breast cancer cases and 25 579 controls. Hum. Mol. Genet. 22, 2539–2550 (2013).
Mogushi, K. & Tanaka, H. PathAct: a novel method for pathway analysis using gene expression profiles. Bioinformation 9, 394–400 (2013).
Chung, R.-H. & Chen, Y.-E. A two-stage random forest-based pathway analysis method. PLoS ONE 7, e36662 (2012).
Bailey, L. R., Roodi, N., Dupont, W. D. & Parl, F. F. Association of cytochrome P450 1B1 (CYP1B1) polymorphism with steroid receptor status in breast cancer. Cancer Res. 58, 5038–5041 (1998).
Shabalin, A. A. Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics 28, 1353–1358 (2012).
Abecasis, G. R., Cardon, L. R. & Cookson, W. O. A general test of association for quantitative traits in nuclear families. Am. J. Hum. Genet. 66, 279–292 (2000).
Rozowsky, J. et al. AlleleSeq: analysis of allele-specific expression and binding in a network framework. Mol. Syst. Biol. 7, 522 (2011).
Wang, K., Li, M. & Hakonarson, H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164 (2010).
Ward, L. D. & Kellis, M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 40, D930–D934 (2012).
Boyle, A. P. et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 22, 1790–1797 (2012).
Emilsson, V. et al. Genetics of gene expression and its effect on disease. Nature 452, 423–428 (2008). This important paper presents the relationship between genetic variation, gene expression and clinical phenotypes using human blood and adipose tissue.
Acknowledgements
Support for the authors was provided by the US National Institutes of Health grants LM010040 (ATHENA) and HL065962 (the P-STAR Network Resource of the PGRN). E.R.H. was funded by grant Z01 HG00153-08-IDRB. R.L. was funded by the US National Science Foundation under Grant number DGE1255832. Any opinions, findings and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the US National Science Foundation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Glossary
- Complex traits
-
Characteristics that arise from interactions among multiple molecular factors, with the potential influence of environmental and behavioural factors. Complex traits do not conform to the inheritance pattern of Mendelian traits.
- Meta-dimensional analysis
-
An approach whereby all scales of data are combined simultaneously to produce complex models defined as multiple variables from multiple scales of data.
- Multi-staged analysis
-
A stepwise or hierarchical analysis method that reduces the search space through different stages of analysis.
- Systems genomics
-
An analysis approach that models the complex inter- and intra-individual variations of traits and diseases using data from next-generation omic data.
- Data integration
-
The incorporation of multi-omic information in a meaningful way to provide a more comprehensive analysis of a biological point of interest.
- Quality control
-
Various techniques used to remove noise and confounding factors from the data.
- Factor analysis
-
A statistical method used to describe variability among observed, correlated variables in terms of a smaller number of unobserved (latent) variables.
- Multi-omics data
-
Multiple types of genome-scale data sets that emerged from high-throughput technologies, including genome sequencing data (genomics), genome-wide RNA-sequencing data (transcriptomics), methylation and histone modification data (epigenomics), and mass spectrometry protein data (proteomics).
- Population stratification
-
A situation in which different subpopulations exist within a data set owing to different allele frequencies because of underlying genetic ancestry that leads to different strata being present in the data set. This can lead to spurious associations if not adjusted for appropriately.
- Multivariate Cox LASSO (least absolute shrinkage and selection operator) model
-
A method that performs variable selection via LASSO, followed by a multivariate Cox regression analysis.
- Kernel-based integration
-
The use of a valid kernel to perform a data matrix transformation before the integration of multiple data types.
- Graph-based integration
-
The use of graphs to perform a data matrix transformation before integration. A graph is a natural method for analysing relationships between samples, as the nodes depict individual samples and the edges represent their possible relationships.
- Majority voting
-
A method in which multiple models are constructed and subsequently evaluated to determine which performs best.
- Ensemble classifiers
-
Classifiers constructed through the use of multiple learning methods to obtain better predictive performance than could be obtained from any of the individual learning algorithms.
- Bayesian network
-
A type of statistical model that represents a set of random variables and their conditional dependencies via a directed acyclic graph.
- Overfitting
-
Building a statistical model that explains the training data set that but does not generalize to independent data.
- Type I errors
-
(Also known as false positives). The acceptance of the alternative hypothesis when the null hypothesis is true.
- Genome-wide association studies
-
Studies that aim to identify disease- or trait-related genetic variations from the whole genome.
Rights and permissions
About this article
Cite this article
Ritchie, M., Holzinger, E., Li, R. et al. Methods of integrating data to uncover genotype–phenotype interactions. Nat Rev Genet 16, 85–97 (2015). https://doi.org/10.1038/nrg3868
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrg3868
This article is cited by
-
Multimodal epigenetic sequencing analysis (MESA) of cell-free DNA for non-invasive colorectal cancer detection
Genome Medicine (2024)
-
Multi-omics assists genomic prediction of maize yield with machine learning approaches
Molecular Breeding (2024)
-
Clonostachys rosea ‘omics profiling: identification of putative metabolite-gene associations mediating its in vitro antagonism against Fusarium graminearum
BMC Genomics (2023)
-
Asterics: a simple tool for the ExploRation and Integration of omiCS data
BMC Bioinformatics (2023)
-
DeeP4med: deep learning for P4 medicine to predict normal and cancer transcriptome in multiple human tissues
BMC Bioinformatics (2023)