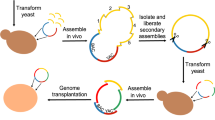
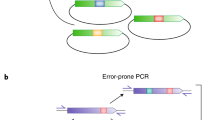
Abstract
Genome engineering strategies — such as genome editing, reduction and shuffling, and de novo genome synthesis — enable the modification of specific genomic locations in a directed and combinatorial manner. These approaches offer an unprecedented opportunity to study central evolutionary issues in which natural genetic variation is limited or biased, which sheds light on the evolutionary forces driving complex and extremely slowly evolving traits; the selective constraints on genome architecture; and the reconstruction of ancestral states of cellular structures and networks.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Barrick, J. E. & Lenski, R. E. Genome dynamics during experimental evolution. Nature Rev. Genet. 14, 827–839 (2013).
Vieira-Silva, S. & Rocha, E. P. The systemic imprint of growth and its uses in ecological (meta)genomics. PLoS Genet. 6, e1000808 (2010).
Elena, S. F. & Lenski, R. E. Evolution experiments with microorganisms: the dynamics and genetic bases of adaptation. Nature Rev. Genet. 4, 457–469 (2003).
Dettman, J. R. et al. Evolutionary insight from whole-genome sequencing of experimentally evolved microbes. Mol. Ecol. 21, 2058–2077 (2012).
Barrick, J. E. et al. Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature 461, 1243–1247 (2009).
Blount, Z. D., Borland, C. Z. & Lenski, R. E. Historical contingency and the evolution of a key innovation in an experimental population of Escherichia coli. Proc. Natl Acad. Sci. USA 105, 7899–7906 (2008).
Nilsson, A. I. et al. Bacterial genome size reduction by experimental evolution. Proc. Natl Acad. Sci. USA 102, 12112–12116 (2005).
Romero, P. A. & Arnold, F. H. Exploring protein fitness landscapes by directed evolution. Nature Rev. Mol. Cell Biol. 10, 866–876 (2009).
Peisajovich, S. G. & Tawfik, D. S. Protein engineers turned evolutionists. Nature Methods 4, 991–994 (2007).
Esvelt, K. M. & Wang, H. H. Genome-scale engineering for systems and synthetic biology. Mol. Systems Biol. 9, 641 (2013).
Carr, P. A. & Church, G. M. Genome engineering. Nature Biotech. 27, 1151–1162 (2009).
Woodruff, L. B. & Gill, R. T. Engineering genomes in multiplex. Curr. Opin. Biotechnol. 22, 576–583 (2011).
Wang, H. H. et al. Programming cells by multiplex genome engineering and accelerated evolution. Nature 460, 894–898 (2009).
Lajoie, M. J. et al. Genomically recoded organisms expand biological functions. Science 342, 357–360 (2013).
Isaacs, F. J. et al. Precise manipulation of chromosomes in vivo enables genome-wide codon replacement. Science 333, 348–353 (2011).
Pósfai, G. et al. Emergent properties of reduced-genome Escherichia coli. Science 312, 1044–1046 (2006).
Patnaik, R. et al. Genome shuffling of Lactobacillus for improved acid tolerance. Nature Biotech. 20, 707–712 (2002).
Zhang, Y. X. et al. Genome shuffling leads to rapid phenotypic improvement in bacteria. Nature 415, 644–646 (2002).
Lartigue, C. et al. Genome transplantation in bacteria: changing one species to another. Science 317, 632–638 (2007).
Rocha, E. P. The organization of the bacterial genome. Annu. Rev. Genet. 42, 211–233 (2008).
Knight, R. D., Freeland, S. J. & Landweber, L. F. Rewiring the keyboard: evolvability of the genetic code. Nature Rev. Genet. 2, 49–58 (2001).
Ambrogelly, A., Palioura, S. & Söll, D. Natural expansion of the genetic code. Nature Chem. Biol. 3, 29–35 (2007).
Lajoie, M. J. et al. Probing the limits of genetic recoding in essential genes. Science 342, 361–363 (2013).
Bezerra, A. R. et al. Reversion of a fungal genetic code alteration links proteome instability with genomic and phenotypic diversification. Proc. Natl Acad. Sci. USA 110, 11079–11084 (2013).
Liu, C. C. & Schultz, P. G. Adding new chemistries to the genetic code. Annu. Rev. Biochem. 79, 413–444 (2010).
Philip, G. K. & Freeland, S. J. Did evolution select a nonrandom “alphabet” of amino acids? Astrobiology 11, 235–240 (2011).
Liu, C. C. et al. Protein evolution with an expanded genetic code. Proc. Natl Acad. Sci. 105, 17688–17693 (2008).
Walter, K. U., Vamvaca, K. & Hilvert, D. An active enzyme constructed from a 9-amino acid alphabet. J. Biol. Chem. 280, 37742–37746 (2005).
Hammerling, M. J. et al. Bacteriophages use an expanded genetic code on evolutionary paths to higher fitness. Nature Chem. Biol. 10, 178 (2014).
Wernegreen, J. J. Genome evolution in bacterial endosymbionts of insects. Nature Rev. Genet. 3, 850–861 (2002).
Fehér, T., Papp, B., Pál, C. & Pósfai, G. Systematic genome reductions: theoretical and experimental approaches. Chem. Rev. 107, 3498–3513 (2007).
Pál, C., Papp, B. & Lercher, M. J. Adaptive evolution of bacterial metabolic networks by horizontal gene transfer. Nature Genet. 37, 1372–1375 (2005).
Umenhoffer, K. et al. Reduced evolvability of Escherichia coli MDS42, an IS-less cellular chassis for molecular and synthetic biology applications. Microb. Cell Fact 9, 38 (2010).
Fehér, T. et al. Competition between transposable elements and mutator genes in bacteria. Mol. Biol. Evol. 29, 3153–3159 (2012).
Pál, C. et al. Chance and necessity in the evolution of minimal metabolic networks. Nature 440, 667–670 (2006).
Hurst, L. D., Pál, C. & Lercher, M. J. The evolutionary dynamics of eukaryotic gene order. Nature Rev. Genet. 5, 299–310 (2004).
Dymond, J. S. et al. Synthetic chromosome arms function in yeast and generate phenotypic diversity by design. Nature 477, 471–476 (2011).
Teresa Avelar, A., Perfeito, L., Gordo, I. & Godinho Ferreira, M. Genome architecture is a selectable trait that can be maintained by antagonistic pleiotropy. Nature Commun. 4, 2235 (2013).
Annaluru, N. et al. Total synthesis of a functional designer eukaryotic chromosome. Science 344, 55–58 (2014).
Lynch, M. & Abegg, A. The rate of establishment of complex adaptations. Mol. Biol. Evol. 27, 1404–1414 (2010).
Nyerges, A. et al. Conditional DNA repair mutants enable highly precise genome engineering. Nucleic Acids Res. 42, e62 (2014).
Warner, J. R., Reeder, P. J., Karimpour-Fard, A., Woodruff, L. B. & Gill, R. T. Rapid profiling of a microbial genome using mixtures of barcoded oligonucleotides. Nature Biotech. 28, 856–862 (2010).
Papp, B., Notebaart, R. A. & Pál, C. Systems-biology approaches for predicting genomic evolution. Nature Rev. Genet. 12, 591–602 (2011).
Wang, Z., Liao, B. Y. & Zhang, J. Genomic patterns of pleiotropy and the evolution of complexity. 107, 18034–18039 (2010).
Benner, S. A., Sassi, S. O. & Gaucher, E. A. Molecular paleoscience: systems biology from the past. Adv. Enzymol. Relat. Areas Mol. Biol. 75, 1–132 (2007).
Voordeckers, K. et al. Reconstruction of ancestral metabolic enzymes reveals molecular mechanisms underlying evolutionary innovation through gene duplication. PLoS Biol. 10, e1001446 (2012).
Finnigan, G. C., Hanson-Smith, V., Stevens, T. H. & Thornton, J. W. Evolution of increased complexity in a molecular machine. Nature 481, 360–364 (2012).
Gaucher, E. A., Thomson, J. M., Burgan, M. F. & Benner, S. A. Inferring the palaeoenvironment of ancient bacteria on the basis of resurrected proteins. Nature 425, 285–288 (2003).
Isalan, M. et al. Evolvability and hierarchy in rewired bacterial gene networks. Nature 452, 840–845 (2008).
Cagatay, T., Turcotte, M., Elowitz, M. B., Garcia-Ojalvo, J. & Süel, G. M. Architecture-dependent noise discriminates functionally analogous differentiation circuits. Cell 139, 512–522 (2009).
Haseltine, E. L. & Arnold, F. H. Synthetic gene circuits: design with directed evolution. Annu. Rev. Biophys. Biomol. Struct. 36, 1–19 (2007).
Peisajovich, S. G., Garbarino, J. E., Wei, P. & Lim, W. A. Rapid diversification of cell signaling phenotypes by modular domain recombination. Science 328, 368–372 (2010).
Yokobayashi, Y., Weiss, R. & Arnold, F. H. Directed evolution of a genetic circuit. Proc. Natl Acad. Sci. USA 99, 16587–16591 (2002).
Renda, B. A., Hammerling, M. J. & Barrick, J. E. Engineering reduced evolutionary potential for synthetic biology. Mol. Biosyst. http://dx.doi.org/10.1039/C3MB70606K (2014).
Fehér, T. et al. Scarless engineering of the Escherichia coli genome. Methods Mol. Biol. 416, 251–259 (2008).
Csörgo, B., Fehér, T., Tímár, E., Blattner, F. R. & Pósfai, G. Low-mutation-rate, reduced-genome Escherichia coli: an improved host for faithful maintenance of engineered genetic constructs. Microb. Cell Fact 11, 11 (2012).
Ellis, H. M., Yu, D., DiTizio, T. & Court, D. L. High efficiency mutagenesis, repair, and engineering of chromosomal DNA using single-stranded oligonucleotides. Proc. Natl Acad. Sci. USA 98, 6742–6746 (2001).
Soskine, M. & Tawfik, D. S. Mutational effects and the evolution of new protein functions. Nature Rev. Genet. 11, 572–582 (2010).
Bogdanove, A. J. & Voytas, D. F. TAL effectors: customizable proteins for DNA targeting. Science 333, 1843–1846 (2011).
Jiang, W., Bikard, D., Cox, D., Zhang, F. & Marraffini, L. A. RNA-guided editing of bacterial genomes using CRISPR–Cas systems. Nature Biotech. 31, 233–239 (2013).
Enyeart, P. J. et al. Generalized bacterial genome editing using mobile group II introns and Cre–lox. Mol. Syst. Biol. 9, 685 (2013).
Gibson, D. G. et al. Creation of a bacterial cell controlled by a chemically synthesized genome. Science 329, 52–56 (2010).
Chan, L. Y., Kosuri, S. & Endy, D. Refactoring bacteriophage T7. Mol. Syst. Biol. 1, 2005.0018 (2005).
Rhodius, V. A. et al. Design of orthogonal genetic switches based on a crosstalk map of σs, anti-σs, and promoters. Mol. Syst. Biol. 9, 702 (2013).
Itaya, M., Tsuge, K., Koizumi, M. & Fujita, K. Combining two genomes in one cell: stable cloning of the Synechocystis PCC6803 genome in the Bacillus subtilis 168 genome. Proc. Natl Acad. Sci. USA 102, 15971–15976 (2005).
Acknowledgements
The authors thank the anonymous reviewers for suggestions on the manuscript. C.P. and B.P. thank the Wellcome Trust and the Lendulet Programme of the Hungarian Academy of Sciences for supporting this work; G.P. thanks the Hungarian Research Council (OTKA) for supporting this work. B. Kintses, A. Nyerges and B. Csorgo gave comments on an earlier version of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Glossary
- Amino acid 'alphabet'
-
The set of amino acids used to build genetically encoded proteins.
- Antagonistic pleiotropy
-
Pleiotropy occurs when a single gene influences multiple phenotypic traits that are seemingly unrelated. In the case of antagonistic pleiotropy, expression of the pleiotropic gene has mixed, competing effects; some of these are beneficial but others are detrimental to the organism.
- Codon ambiguity
-
An extreme form of mistranslation in which a codon can be translated as two different amino acids.
- Combinatorial explosion
-
A fundamental problem in evolutionary optimization and computing. As the size of the investigated system and the number of corresponding parameters increase, the number of combinations that one has to examine grows exponentially, which requires an intolerable amount of time to examine them.
- Convergent evolution
-
Evolution of similar phenotypes in different populations or species as a result of adaptation to similar environments or ecological niches.
- Directed protein evolution
-
A protein engineering method to evolve proteins with desirable properties. It mimics and accelerates natural evolutionary processes by applying in vitro diversification–selection–amplification cycles.
- Epistatic interactions
-
Interactions between two mutations whereby the phenotypic effect of one mutation depends on the presence of another mutation.
- Genome editing
-
Modification of the genetic information encoded by the genome using in vivo, directed modification (such as replacement, removal or insertion of DNA bases) of a single locus or multiple loci. It uses synthetic oligonucleotides and a range of accessory tools, including engineered nucleases, and DNA repair and recombination enzymes.
- Leading DNA strand
-
The strand of nascent DNA that is being 'read' by the DNA polymerase in the same direction as the replication fork proceeds. It is being synthesized continuously, as opposed to the lagging strand.
- Minimal genomes
-
Genomes that carry only the minimal genetic information necessary for life in a given environmental condition. Reduction towards a minimal essential gene set can occur either naturally (for example, in symbionts) or by genetic engineering.
- Multiplex automated genome engineering
-
(MAGE). A highly efficient genome editing method that can generate a large and heterogeneous population of mutant bacterial genomes within days. Using oligonucleotide-mediated allelic replacement technology in a cyclic and automated manner, MAGE can simultaneously target and modify multiple genomic locations across a large population of cells.
- Site-specific recombineering
-
A recombination engineering system that allows efficient manipulation of genomic DNA at predetermined locations. It does not require extensive sequence similarity and relies on site-specific recombinases that catalyse reciprocal recombination of DNA at short sequences.
- Synthetic chromosome
-
An artificial chromosome synthesized from simple chemical building blocks. Owing to limitations in the length of DNA that is amenable to direct chemical synthesis, construction of synthetic chromosomes is a hierarchical process, in which synthetic oligonucleotides are assembled into larger DNA segments in a step-wise manner using in vitro and in vivo assembly methods.
Rights and permissions
About this article
Cite this article
Pál, C., Papp, B. & Pósfai, G. The dawn of evolutionary genome engineering. Nat Rev Genet 15, 504–512 (2014). https://doi.org/10.1038/nrg3746
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrg3746
This article is cited by
-
A novel method for achieving an optimal classification of the proteinogenic amino acids
Scientific Reports (2020)
-
Development of a high-copy-number plasmid via adaptive laboratory evolution of Corynebacterium glutamicum
Applied Microbiology and Biotechnology (2018)
-
An evolutionary optimization of a rhodopsin-based phototrophic metabolism in Escherichia coli
Microbial Cell Factories (2017)
-
Genome reprogramming for synthetic biology
Frontiers of Chemical Science and Engineering (2017)
-
Genomes by design
Nature Reviews Genetics (2015)