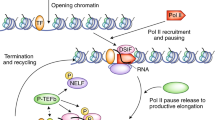
Key Points
-
Transcriptional regulation is highly complex on many timescales.
-
The binding of transcription factors to eukaryotic chromosomes is strongly restricted by complex chromatin structures.
-
Through the recruitment of chromatin-modifying activities that reorganize local nucleosome structures, transcription factors gain access to regulatory elements.
-
Genome-wide methodologies provide extensive data sets on the localization of transcription factors, global histone modifications and regions of accessible chromatin.
-
By their nature, these approaches average signals across large cell populations that represent many heterogeneous states at the molecular level and lead to a static view of transcriptional states.
-
By contrast, single-cell approaches have shown that many of the molecular factor–template interactions are highly dynamic in living cells.
-
These dynamic processes are integrated over the lifetime of the cell cycle to create transcriptional profiles that often fluctuate with time, either in response to external signals or as a direct consequence of the internal molecular networks.
-
A rigorous mechanistic understanding of fundamental transcriptional processes must integrate both the large-scale identification of factors and their activities, and the highly dynamic molecular processes that underlie their function.
Abstract
The interaction of regulatory proteins with the complex nucleoprotein structures that are found in mammalian cells involves chromatin reorganization at multiple levels. Mechanisms that support these transitions are complex on many timescales, which range from milliseconds to minutes or hours. In this Review, we discuss emerging concepts regarding the function of regulatory elements in living cells. We also explore the involvement of these dynamic and stochastic processes in the evolution of fluctuating transcriptional activity states that are now commonly reported in eukaryotic systems.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Johnson, D. G. & Dent, S. Y. Chromatin: receiver and quarterback for cellular signals. Cell 152, 685–689 (2013).
Hardison, R. C. & Taylor, J. Genomic approaches towards finding cis-regulatory modules in animals. Nature Rev. Genet. 13, 469–483 (2012).
Baylin, S. B. & Jones, P. A. A decade of exploring the cancer epigenome — biological and translational implications. Nature Rev. Cancer 11, 726–734 (2011).
Spitz, F. & Furlong, E. E. Transcription factors: from enhancer binding to developmental control. Nature Rev. Genet. 13, 613–626 (2012).
Wu, W. et al. Dynamics of the epigenetic landscape during erythroid differentiation after GATA1 restoration. Genome Res. 21, 1659–1671 (2011).
Reik, W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature 447, 425–432 (2007).
Spivakov, M. & Fisher, A. G. Epigenetic signatures of stem-cell identity. Nature Rev. Genet. 8, 263–271 (2007).
Rivera, C. M. & Ren, B. Mapping human epigenomes. Cell 155, 39–55 (2013).
Thurman, R. E. et al. The accessible chromatin landscape of the human genome. Nature 489, 75–82 (2012).
Degner, J. F. et al. DNase I sensitivity QTLs are a major determinant of human expression variation. Nature 482, 390–394 (2012).
Sanyal, A., Lajoie, B. R., Jain, G. & Dekker, J. The long-range interaction landscape of gene promoters. Nature 489, 109–113 (2012).
Larson, D. R. What do expression dynamics tell us about the mechanism of transcription? Curr. Opin. Genet. Dev. 21, 591–599 (2011).
Mellor, J. Dynamic nucleosomes and gene transcription. Trends Genet. 22, 320–329 (2006).
McNally, J. G., Mueller, W. G., Walker, D., Wolford, R. G. & Hager, G. L. The glucocorticoid receptor: rapid exchange with regulatory sites in living cells. Science 287, 1262–1265 (2000). This paper reports the first observation of site-specific factor binding to a regulatory element in living cells, which reveals rapid exchange dynamics.
Kaern, M., Elston, T. C., Blake, W. J. & Collins, J. J. Stochasticity in gene expression: from theories to phenotypes. Nature Rev. Genet. 6, 451–464 (2005).
Coulon, A., Chow, C. C., Singer, R. H. & Larson, D. R. Eukaryotic transcriptional dynamics: from single molecules to cell populations. Nature Rev. Genet. 14, 572–584 (2013).
Boyle, A. P. et al. High-resolution mapping and characterization of open chromatin across the genome. Cell 132, 311–322 (2008).
John, S. et al. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nature Genet. 43, 264–268 (2011).
Hoogenkamp, M. et al. Early chromatin unfolding by RUNX1: a molecular explanation for differential requirements during specification versus maintenance of the hematopoietic gene expression program. Blood 114, 299–309 (2009).
Siersbaek, R. et al. Extensive chromatin remodelling and establishment of transcription factor 'hotspots' during early adipogenesis. EMBO J. 30, 1459–1472 (2011).
Stamatoyannopoulos, J. A. et al. An encyclopedia of mouse DNA elements (mouse ENCODE). Genome Biol. 13, 418 (2012).
Biddie, S. C. et al. Transcription factor AP1 potentiates chromatin accessibility and glucocorticoid receptor binding. Mol. Cell 43, 145–155 (2011).
Heintzman, N. D. & Ren, B. Finding distal regulatory elements in the human genome. Curr. Opin. Genet. Dev. 19, 541–549 (2009).
Bosisio, D. et al. A hyper-dynamic equilibrium between promoter-bound and nucleoplasmic dimers controls NF-κB-dependent gene activity. EMBO J. 25, 798–810 (2006).
Sharp, Z. D. et al. Estrogen-receptor-α exchange and chromatin dynamics are ligand-and domain-dependent. J. Cell Sci. 119, 4101–4116 (2006).
Yao, J., Munson, K. M., Webb, W. W. & Lis, J. T. Dynamics of heat shock factor association with native gene loci in living cells. Nature 442, 1050–1053 (2006).
Lickwar, C. R., Mueller, F., Hanlon, S. E., McNally, J. G. & Lieb, J. D. Genome-wide protein–DNA binding dynamics suggest a molecular clutch for transcription factor function. Nature 484, 251–255 (2012).
Kuipers, M. A. et al. Highly stable loading of Mcm proteins onto chromatin in living cells requires replication to unload. J. Cell Biol. 192, 29–41 (2011).
Stasevich, T. J. & McNally, J. G. Assembly of the transcription machinery: ordered and stable, random and dynamic, or both? Chromosoma 120, 533–545 (2011).
Mueller, F., Mazza, D., Stasevich, T. J. & McNally, J. G. FRAP and kinetic modeling in the analysis of nuclear protein dynamics: what do we really know? Curr. Opin. Cell Biol. 22, 403–411 (2010).
Neph, S. et al. An expansive human regulatory lexicon encoded in transcription factor footprints. Nature 489, 83–90 (2012). This study carries out an exhaustive characterization of DNaseI hypersensitive regions in human cell lines, which reveals the presence of many transcription factors by their footprints within the accessible region.
Deng, W. et al. Controlling long-range genomic interactions at a native locus by targeted tethering of a looping factor. Cell 149, 1233–1244 (2012). This study finds that LIM domain-binding protein 1(LDB1)effects GATA1-mediated chromatin loop formation by long-range protein–protein interactions.
Stender, J. D. et al. Genome-wide analysis of estrogen receptor α DNA binding and tethering mechanisms identifies Runx1 as a novel tethering factor in receptor-mediated transcriptional activation. Mol. Cell. Biol. 30, 3943–3955 (2010).
Erdel, F., Krug, J., Langst, G. & Rippe, K. Targeting chromatin remodelers: signals and search mechanisms. Biochim. Biophys. Acta 1809, 497–508 (2011).
Cairns, B. R. Chromatin remodeling: insights and intrigue from single-molecule studies. Nature Struct. Mol. Biol. 14, 989–996 (2007).
Clapier, C. R. & Cairns, B. R. The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 78, 273–304 (2009).
Glatt, S., Alfieri, C. & Muller, C. W. Recognizing and remodeling the nucleosome. Curr. Opin. Struct. Biol. 21, 335–341 (2011).
Hargreaves, D. C. & Crabtree, G. R. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res. 21, 396–420 (2011).
Korber, P. & Becker, P. B. Nucleosome dynamics and epigenetic stability. Essays Biochem. 48, 63–74 (2010).
Miller, J. A. & Widom, J. Collaborative competition mechanism for gene activation in vivo. Mol. Cell. Biol. 23, 1623–1632 (2003).
Mirny, L. A. Nucleosome-mediated cooperativity between transcription factors. Proc. Natl Acad. Sci. USA 107, 22534–22539 (2010).
Teif, V. B., Ettig, R. & Rippe, K. A lattice model for transcription factor access to nucleosomal DNA. Biophys. J. 99, 2597–2607 (2010).
Zaret, K. S. & Carroll, J. S. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 25, 2227–2241 (2011).
Hatta, M. & Cirillo, L. A. Chromatin opening and stable perturbation of core histone: DNA contacts by FoxO1. J. Biol. Chem. 282, 35583–35593 (2007).
Magnani, L., Eeckhoute, J. & Lupien, M. Pioneer factors: directing transcriptional regulators within the chromatin environment. Trends Genet. 27, 465–474 (2011).
Lupien, M. et al. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell 132, 958–970 (2008).
Jozwik, K. M. & Carroll, J. S. Pioneer factors in hormone-dependent cancers. Nature Rev. Cancer 12, 381–385 (2012).
Cirillo, L. A. & Zaret, K. S. Specific interactions of the wing domains of FOXA1 transcription factor with DNA. J. Mol. Biol. 366, 720–724 (2007). In this study, FOXA1 is characterized as a pioneer protein through site-specific interactions with DNA.
Boeger, H., Griesenbeck, J., Strattan, J. S. & Kornberg, R. D. Removal of promoter nucleosomes by disassembly rather than sliding in vivo. Mol. Cell 14, 667–673 (2004).
Boeger, H., Griesenbeck, J. & Kornberg, R. D. Nucleosome retention and the stochastic nature of promoter chromatin remodeling for transcription. Cell 133, 716–726 (2008). This paper presents nucleosome occupancy at the yeast PHO5 locus as a dynamic equilibrium between multiple states.
Nagaich, A. K., Walker, D. A., Wolford, R. G. & Hager, G. L. Rapid periodic binding and displacement of the glucocorticoid receptor during chromatin remodeling. Mol. Cell 14, 163–174 (2004). This study uses UV laser crosslinking to characterize dynamics of factor binding during chromatin remodelling.
McKnight, J. N., Jenkins, K. R., Nodelman, I. M., Escobar, T. & Bowman, G. D. Extranucleosomal DNA binding directs nucleosome sliding by chd1. Mol. Cell. Biol. 31, 4746–4759 (2011). This paper shows that nucleosome invasion of a factor-specific binding site, which is catalysed by a remodelling protein, displaces the binding complex from the site.
Kassabov, S. R., Henry, N. M., Zofall, M., Tsukiyama, T. & Bartholomew, B. High-resolution mapping of changes in histone–DNA contacts of nucleosomes remodeled by ISW2. Mol. Cell. Biol. 22, 7524–7534 (2002).
Voss, T. C. et al. Dynamic exchange at regulatory elements during chromatin remodeling underlies assisted loading mechanism. Cell 146, 544–554 (2011). This study shows that two factors that bind to the same DNA recognition element fail to manifest competition in living cells, which leads to the hypothesis of dynamic assisted loading.
MacArthur, S. et al. Developmental roles of 21 Drosophila transcription factors are determined by quantitative differences in binding to an overlapping set of thousands of genomic regions. Genome Biol. 10, R80 (2009).
Moorman, C. et al. Hotspots of transcription factor colocalization in the genome of Drosophila melanogaster. Proc. Natl Acad. Sci. USA 103, 12027–12032 (2006). This paper shows that the Drosophila melanogaster genome contains many hot spots that are targeted by multiple transcription factors.
Zinzen, R. P., Girardot, C., Gagneur, J., Braun, M. & Furlong, E. E. Combinatorial binding predicts spatio-temporal cis-regulatory activity. Nature 462, 65–70 (2009).
Farnham, P. J. Insights from genomic profiling of transcription factors. Nature Rev. Genet. 10, 605–616 (2009).
modENCODE Consortium et al. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science 330, 1787–1797 (2010).
Grandori, C., Cowley, S. M., James, L. P. & Eisenman, R. N. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu. Rev. Cell Dev. Biol. 16, 653–699 (2000).
Rigaud, G., Roux, J., Pictet, R. & Grange, T. In vivo footprinting of rat TAT gene: dynamic interplay between the glucocorticoid receptor and a liver-specific factor. Cell 67, 977–986 (1991). This paper shows that the glucocorticoid receptor induces binding of a liver transcription factor upstream of the tyrosine aminotransferase gene ( Tat ), even though the receptor and the liver factor compete for binding as pure proteins on naked DNA.
Morris, S. A. et al. Overlapping chromatin-remodeling systems collaborate genome wide at dynamic chromatin transitions. Nature Struct. Mol. Biol. http://dx.doi.org/10.1038/nsmb.2718 (2013).
Hilfinger, A. & Paulsson, J. Separating intrinsic from extrinsic fluctuations in dynamic biological systems. Proc. Natl Acad. Sci. USA 108, 12167–12172 (2011).
Swain, P. S., Elowitz, M. B. & Siggia, E. D. Intrinsic and extrinsic contributions to stochasticity in gene expression. Proc. Natl Acad. Sci. USA 99, 12795–12800 (2002).
Harper, C. V. et al. Dynamic analysis of stochastic transcription cycles. PLoS. Biol. 9, e1000607 (2011).
John, S. et al. Kinetic complexity of the global response to glucocorticoid receptor action. Endocrinology 150, 1766–1774 (2009).
Voss, T. C., John, S. & Hager, G. L. Single cell analysis of glucocorticoid receptor action reveals that stochastic post-chromatin association mechanisms regulate ligand-specific transcription. Mol. Endocrinol. 20, 2641–2655 (2006).
Elowitz, M. B., Levine, A. J., Siggia, E. D. & Swain, P. S. Stochastic gene expression in a single cell. Science 297, 1183–1186 (2002).
Raser, J. M. & O'Shea, E. K. Control of stochasticity in eukaryotic gene expression. Science 304, 1811–1814 (2004).
Alon, U. Network motifs: theory and experimental approaches. Nature Rev. Genet. 8, 450–461 (2007).
Yosef, N. & Regev, A. Impulse control: temporal dynamics in gene transcription. Cell 144, 886–896 (2011).
Veldhuis, J. D., Keenan, D. M. & Pincus, S. M. Motivations and methods for analyzing pulsatile hormone secretion. Endocr. Rev. 29, 823–864 (2008).
Stavreva, D. A. et al. Ultradian hormone stimulation induces glucocorticoid receptor-mediated pulses of gene transcription. Nature Cell Biol. 11, 1093–1102 (2009).
Stavreva, D. A., Varticovski, L. & Hager, G. L. Complex dynamics of transcription regulation. Biochim. Biophys. Acta 1819, 657–666 (2012).
Hughes, M. E. et al. Harmonics of circadian gene transcription in mammals. PLoS. Genet. 5, e1000442 (2009).
Vollmers, C. et al. Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression. Proc. Natl Acad. Sci. USA 106, 21453–21458 (2009).
Ou, Q., Magico, A. & King-Jones, K. Nuclear receptor DHR4 controls the timing of steroid hormone pulses during Drosophila development. PLoS. Biol. 9, e1001160 (2011).
Uhlendorf, J. et al. Long-term model predictive control of gene expression at the population and single-cell levels. Proc. Natl Acad. Sci. USA 109, 14271–14276 (2012).
Hoffmann, A. & Baltimore, D. Circuitry of nuclear factor κB signaling. Immunol. Rev. 210, 171–186 (2006).
Sung, M. H. et al. Sustained oscillations of NF-κB produce distinct genome scanning and gene expression profiles. PLoS ONE 4, e7163 (2009).
Nelson, D. E. et al. Oscillations in NF-κB signaling control the dynamics of gene expression. Science 306, 704–708 (2004).
Lahav, G. et al. Dynamics of the p53–Mdm2 feedback loop in individual cells. Nature Genet. 36, 147–150 (2004). This study shows that p53 is expressed in discrete pulses after DNA damage and that the number, but not the size, of pulses increases with the extent of DNA damage.
Ashall, L. et al. Pulsatile stimulation determines timing and specificity of NF-κB-dependent transcription. Science 324, 242–246 (2009).
Purvis, J. E. et al. p53 dynamics control cell fate. Science 336, 1440–1444 (2012).
Bratsun, D., Volfson, D., Tsimring, L. S. & Hasty, J. Delay-induced stochastic oscillations in gene regulation. Proc. Natl Acad. Sci. USA 102, 14593–14598 (2005).
Cai, L., Dalal, C. K. & Elowitz, M. B. Frequency-modulated nuclear localization bursts coordinate gene regulation. Nature 455, 485–490 (2008).
Hao, N. & O'Shea, E. K. Signal-dependent dynamics of transcription factor translocation controls gene expression. Nature Struct. Mol. Biol. 19, 31–39 (2012).
Blake, W. J., Kaern, M., Cantor, C. R. & Collins, J. J. Noise in eukaryotic gene expression. Nature 422, 633–637 (2003).
Janicki, S. M. et al. From silencing to gene expression: real-time analysis in single cells. Cell 116, 683–698 (2004).
Golding, I., Paulsson, J., Zawilski, S. M. & Cox, E. C. Real-time kinetics of gene activity in individual bacteria. Cell 123, 1025–1036 (2005).
Chubb, J. R., Trcek, T., Shenoy, S. M. & Singer, R. H. Transcriptional pulsing of a developmental gene. Curr. Biol. 16, 1018–1025 (2006).
Raj, A., Peskin, C. S., Tranchina, D., Vargas, D. Y. & Tyagi, S. Stochastic mRNA synthesis in mammalian cells. PLoS. Biol. 4, e309 (2006).
Darzacq, X. et al. In vivo dynamics of RNA polymerase II transcription. Nature Struct. Mol. Biol. 14, 796–806 (2007).
Suter, D. M., Molina, N., Naef, F. & Schibler, U. Origins and consequences of transcriptional discontinuity. Curr. Opin. Cell Biol. 23, 657–662 (2011).
Wilkinson, D. J. Stochastic modelling for quantitative description of heterogeneous biological systems. Nature Rev. Genet. 10, 122–133 (2009).
Kim, H. D., Shay, T., O'Shea, E. K. & Regev, A. Transcriptional regulatory circuits: predicting numbers from alphabets. Science 325, 429–432 (2009).
Ko, M. S., Nakauchi, H. & Takahashi, N. The dose dependence of glucocorticoid-inducible gene expression results from changes in the number of transcriptionally active templates. EMBO J. 9, 2835–2842 (1990). This paper shows that induction of gene expression by the glucocorticoid receptor results from increases in the frequency of active templates but not from the extent of transcription from each template.
Archer, T. K. et al. Differential steroid hormone induction of transcription from the mouse mammary tumor virus promoter. Mol. Endocrinol. 8, 568–576 (1994).
Becker, M. et al. Dynamic behavior of transcription factors on a natural promoter in living cells. EMBO Rep. 3, 1188–1194 (2002).
Shang, Y., Hu, X., DiRenzo, J., Lazar, M. A. & Brown, M. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell 103, 843–852 (2000).
Metivier, R. et al. Estrogen receptor-α directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell 115, 751–763 (2003).
Hah, N. et al. A rapid, extensive, and transient transcriptional response to estrogen signaling in breast cancer cells. Cell 145, 622–634 (2011).
Karpova, T. S. et al. Concurrent fast and slow cycling of a transcriptional activator at an endogenous promoter. Science 319, 466–469 (2008).
Magklara, A. et al. An epigenetic signature for monoallelic olfactory receptor expression. Cell 145, 555–570 (2011).
Suter, D. M. et al. Mammalian genes are transcribed with widely different bursting kinetics. Science 332, 472–474 (2011). This study expresses a luciferase protein that is detected by bioluminescence in single cells, which reveals bursting kinetics that is characterized by refractory and active periods.
Voss, T. C. et al. Combinatorial probabilistic chromatin interactions produce transcriptional heterogeneity. J. Cell. Sci. 122, 345–356 (2009).
McFerran, D. W. et al. Persistent synchronized oscillations in prolactin gene promoter activity in living pituitary cells. Endocrinology 142, 3255–3260 (2001).
Shorte, S. L. et al. PRL gene expression in individual living mammotropes displays distinct functional pulses that oscillate in a noncircadian temporal pattern. Endocrinology 143, 1126–1133 (2002).
Berno, V. et al. Activation of estrogen receptor-α by E2 or EGF induces temporally distinct patterns of large-scale chromatin modification and mRNA transcription. PLoS ONE. 3, e2286 (2008).
So, L. H. et al. General properties of transcriptional time series in Escherichia coli. Nature Genet. 43, 554–560 (2011).
Mortensen, K. I., Churchman, L. S., Spudich, J. A. & Flyvbjerg, H. Optimized localization analysis for single-molecule tracking and super-resolution microscopy. Nature Methods 7, 377–381 (2010).
Brown, C. M. et al. Raster image correlation spectroscopy (RICS) for measuring fast protein dynamics and concentrations with a commercial laser scanning confocal microscope. J. Microsc. 229, 78–91 (2008).
Digman, M. A., Dalal, R., Horwitz, A. F. & Gratton, E. Mapping the number of molecules and brightness in the laser scanning microscope. Biophys. J. 94, 2320–2332 (2008).
Sengupta, P. et al. Probing protein heterogeneity in the plasma membrane using PALM and pair correlation analysis. Nature Methods 8, 969–975 (2011).
Robertson, G. et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nature Methods 4, 651–657 (2007).
Lickwar, C. R., Mueller, F. & Lieb, J. D. Genome-wide measurement of protein–DNA binding dynamics using competition ChIP. Nature Protoc. 8, 1337–1353 (2013).
Hockensmith, J. W., Kubasek, W. L., Vorachek, W. R., Evertsz, E. M. & von Hippel, P. H. Laser cross-linking of protein–nucleic acid complexes. Methods Enzymol. 208, 211–236 (1991).
Nagaich, A. K. & Hager, G. L. UV laser cross-linking: a real-time assay to study dynamic protein/DNA interactions during chromatin remodeling. Sci. STKE 256, L13 (2004).
Becker, P. B., Ruppert, S. & Schutz, G. Genomic footprinting reveals cell type-specific DNA binding of ubiquitous factors. Cell 51, 435–443 (1987).
Koster, M., Frahm, T. & Hauser, H. Nucleocytoplasmic shuttling revealed by FRAP and FLIP technologies. Curr. Opin. Biotechnol. 16, 28–34 (2005).
Mazza, D., Abernathy, A., Golob, N., Morisaki, T. & McNally, J. G. A benchmark for chromatin binding measurements in live cells. Nucleic Acids Res. 40, e119 (2012).
Sun, Y., Hays, N. M., Periasamy, A., Davidson, M. W. & Day, R. N. Monitoring protein interactions in living cells with fluorescence lifetime imaging microscopy. Methods Enzymol. 504, 371–391 (2012).
Mazza, D., Ganguly, S. & McNally, J. G. Monitoring dynamic binding of chromatin proteins in vivo by single-molecule tracking. Methods Mol. Biol. 1042, 117–137 (2013).
Bustamante, C., Cheng, W. & Mejia, Y. X. Revisiting the central dogma one molecule at a time. Cell 144, 480–497 (2011).
Acknowledgements
The authors acknowledge careful reading of the manuscript and numerous suggestions by L. Grontved and D. Pressman, the US National Institutes of Health.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
FURTHER INFORMATION
Glossary
- DNase hypersensitive sites
-
Local regions of nucleosome reorganization that are detected by their increased accessibility to nucleases.
- Fluorescence recovery after photobleaching
-
(FRAP). An optical technique to measure interaction lifetimes of molecular species. It involves labelling a specific cell component with a fluorescent molecule, followed by photobleaching a sharply defined region of the cell. Imaging is used to observe the subsequent rates and patterns of fluorescence recovery.
- Fluorescence lifetime imaging
-
(FLIM). An alternative fluorescence method to measure lifetimes of molecular species.
- Fluorescence correlation spectroscopy
-
(FCS). A fluorescence method to determine the average lifetime of the interaction between two molecular species when they are present within a given volume.
- Single-molecule tracking
-
A method in which the path of a protein that is labelled with a bright chromophore is followed through the cell in real time.
- Tethering
-
The localization of a protein to a specific site on DNA not by direct DNA binding but by interactions with another protein factor that is bound to the DNA; it is usually detected by chromatin immunoprecipitation.
- ATP-dependent remodelling systems
-
Large multisubunit molecular machines that use ATP energy to reorganize nucleosome structures, often by sliding the nucleosome to a new position on the DNA.
- Pioneer proteins
-
Factors that are proposed to have properties which allow them to penetrate local nucleosome structures and thus to pioneer the recruitment of secondary factors.
- DMS footprinting
-
A method that detects regions of DNA resistance to chemical attack by dimethyl sulphoxide (DMS) owing to the presence of an interacting DNA-binding protein.
- UV laser crosslinking
-
A method that uses dense pulses of high-energy ultraviolet (UV) photons to crosslink DNA bound proteins to the template rapidly and with high efficiency.
- Assisted loading
-
A mechanism that proposes the transient recruitment of a transcription factor to the template by dynamic nucleosome remodelling.
- Transcriptional bursting
-
The phenomenon whereby transcripts are sometimes released from activated promoters as rapid short pulses rather than in a continuous mode.
- Hit-and-run model
-
The hypothesis that many transcription factors reside on specific binding sites for brief periods, which is followed by many of these factors returning to template cycles.
Rights and permissions
About this article
Cite this article
Voss, T., Hager, G. Dynamic regulation of transcriptional states by chromatin and transcription factors. Nat Rev Genet 15, 69–81 (2014). https://doi.org/10.1038/nrg3623
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrg3623