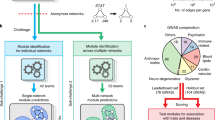
Key Points
-
Bioinformatics approaches for integrating molecular networks across various types of interaction data, omics profiles, conditions or species have demonstrated considerable power for the detection and interpretation of biological modules.
-
Module-discovery approaches are broadly classified into four categories: identification of 'active modules' through the integration of networks and molecular profiles, identification of 'conserved modules' across multiple species, identification of 'differential modules' across different conditions and identification of 'composite modules' through the integration of different interaction types.
-
Active modules mark regions of a network that are most active during a given cellular or disease response and can identify important biomarkers, disease mechanisms and therapeutic targets.
-
Conserved modules are revealed through the alignment or comparison of networks across multiple species. Such modules reflect biologically important pathways that have been conserved over long evolutionary periods.
-
Differential modules are identified through differential analyses of experimentally mapped interactions across multiple conditions.
-
Composite modules are detected through the simultaneous integration of diverse types of molecular interactions.
-
Such integrative approaches reviewed here substantially increase the scope, scale and depth of bioinformatics analysis, by permitting joint interpretation of ensembles of distinct biological information.
Abstract
A central goal of systems biology is to elucidate the structural and functional architecture of the cell. To this end, large and complex networks of molecular interactions are being rapidly generated for humans and model organisms. A recent focus of bioinformatics research has been to integrate these networks with each other and with diverse molecular profiles to identify sets of molecules and interactions that participate in a common biological function — that is, 'modules'. Here, we classify such integrative approaches into four broad categories, describe their bioinformatic principles and review their applications.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Hartwell, L. H., Hopfield, J. J., Leibler, S. & Murray, A. W. From molecular to modular cell biology. Nature 402, C47–C52 (1999).
Alon, U. Biological networks: the tinkerer as an engineer. Science 301, 1866–1867 (2003).
Stuart, J. M., Segal, E., Koller, D. & Kim, S. K. A gene-coexpression network for global discovery of conserved genetic modules. Science 302, 249–255 (2003).
Segal, E. et al. Module networks: identifying regulatory modules and their condition-specific regulators from gene expression data. Nature Genet. 34, 166–176 (2003).
Barabasi, A. L., Gulbahce, N. & Loscalzo, J. Network medicine: a network-based approach to human disease. Nature Rev. Genet. 12, 56–68 (2011).
Barabasi, A. L. & Oltvai, Z. N. Network biology: understanding the cell's functional organization. Nature Rev. Genet. 5, 101–113 (2004).
Spirin, V. & Mirny, L. A. Protein complexes and functional modules in molecular networks. Proc. Natl Acad. Sci. USA 100, 12123–12128 (2003).
Ito, T. et al. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl Acad. Sci. USA 98, 4569–4574 (2001).
Stelzl, U. et al. A human protein-protein interaction network: a resource for annotating the proteome. Cell 122, 957–968 (2005).
Harbison, C. T. et al. Transcriptional regulatory code of a eukaryotic genome. Nature 431, 99–104 (2004).
Ravasi, T. et al. An atlas of combinatorial transcriptional regulation in mouse and man. Cell 140, 744–752 (2010).
Costanzo, M. et al. The genetic landscape of a cell. Science 327, 425–431 (2010).
Collins, S. R. et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature 446, 806–810 (2007).
Rual, J. F. et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature 437, 1173–1178 (2005).
Yu, H. et al. High-quality binary protein interaction map of the yeast interactome network. Science 322, 104–110 (2008).
Muers, M. Systems biology: plant networks. Nature Rev. Genet. 12, 586 (2011).
Milo, R. et al. Network motifs: simple building blocks of complex networks. Science 298, 824–827 (2002).
Ideker, T. & Sharan, R. Protein networks in disease. Genome Res. 18, 644–652 (2008).
Koyuturk, M. Algorithmic and analytical methods in network biology. Wiley Interdiscip. Rev. Syst. Biol. Med. 2, 277–292 (2010).
Fields, S. High-throughput two-hybrid analysis. The promise and the peril. FEBS J. 272, 5391–5399 (2005).
Phizicky, E. M. & Fields, S. Protein-protein interactions: methods for detection and analysis. Microbiol. Rev. 59, 94–123 (1995).
Ben-Hur, A. & Noble, W. S. Kernel methods for predicting protein-protein interactions. Bioinformatics 21 (Suppl. 1), i38–i46 (2005).
Huang, H., Jedynak, B. M. & Bader, J. S. Where have all the interactions gone? Estimating the coverage of two-hybrid protein interaction maps. PLoS Comput. Biol. 3, e214 (2007).
Venkatesan, K. et al. An empirical framework for binary interactome mapping. Nature Methods 6, 83–90 (2009). A critical discussion of biases in high-throughput data analyses that contribute to false positives and negative interpretations.
Cusick, M. E. et al. Literature-curated protein interaction datasets. Nature Methods 6, 39–46 (2009).
Edwards, A. M. et al. Too many roads not taken. Nature 470, 163–165 (2011).
Bandyopadhyay, S. et al. Rewiring of genetic networks in response to DNA damage. Science 330, 1385–1389 (2010). An approach for differential analysis of genetic networks. It was applied to the mapping of DNA damage response pathways in yeast.
Califano, A. Rewiring makes the difference. Mol. Syst. Biol. 7, 463 (2011).
Ideker, T & Bandyopadhyay, S. Integrative systems biology poster [online]. Nature Genet. 42 (2010).
Jenssen, T. K., Laegreid, A., Komorowski, J. & Hovig, E. A literature network of human genes for high-throughput analysis of gene expression. Nature Genet. 28, 21–28 (2001).
Jansen, R., Greenbaum, D. & Gerstein, M. Relating whole-genome expression data with protein-protein interactions. Genome Res. 12, 37–46 (2002).
de Lichtenberg, U., Jensen, L. J., Brunak, S. & Bork, P. Dynamic complex formation during the yeast cell cycle. Science 307, 724–727 (2005).
Segal, E., Wang, H. & Koller, D. Discovering molecular pathways from protein interaction and gene expression data. Bioinformatics 19 (Suppl. 1), i264–i271 (2003).
Gunsalus, K. C. et al. Predictive models of molecular machines involved in Caenorhabditis elegans early embryogenesis. Nature 436, 861–865 (2005).
Jensen, L. J., Jensen, T. S., de Lichtenberg, U., Brunak, S. & Bork, P. Co-evolution of transcriptional and post-translational cell-cycle regulation. Nature 443, 594–597 (2006).
Chen, R. et al. Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell 148, 1293–1307 (2012).
Leiserson, M. D., Blokh, D., Sharan, R. & Raphael, B. J. Simultaneous identification of multiple driver pathways in cancer. PLoS Comput. Biol. 9, e1003054 (2013).
Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455, 1061–1068 (2008).
Nibbe, R. K., Koyuturk, M. & Chance, M. R. An integrative -omics approach to identify functional sub-networks in human colorectal cancer. PLoS Comput. Biol. 6, e1000639 (2010).
Begley, T. J., Rosenbach, A. S., Ideker, T. & Samson, L. D. Hot spots for modulating toxicity identified by genomic phenotyping and localization mapping. Mol. Cell 16, 117–125 (2004).
Guo, Z. et al. Edge-based scoring and searching method for identifying condition-responsive protein-protein interaction sub-network. Bioinformatics 23, 2121–2128 (2007).
Gu, J., Chen, Y., Li, S. & Li, Y. Identification of responsive gene modules by network-based gene clustering and extending: application to inflammation and angiogenesis. BMC Syst. Biol. 4, 47 (2010).
Wu, Z., Zhao, X. & Chen, L. Identifying responsive functional modules from protein-protein interaction network. Mol. Cells 27, 271–277 (2009).
Dittrich, M. T., Klau, G. W., Rosenwald, A., Dandekar, T. & Muller, T. Identifying functional modules in protein-protein interaction networks: an integrated exact approach. Bioinformatics 24, i223–i231 (2008). This article describes a programmatically efficient scheme for module detection that bypasses inherent computational complexities underlying the extraction of high-confidence (that is, maximally scoring) subnetworks from omics data.
Qiu, Y. Q., Zhang, S., Zhang, X. S. & Chen, L. Detecting disease associated modules and prioritizing active genes based on high throughput data. BMC Bioinformatics 11, 26 (2010).
Prelic, A. et al. A systematic comparison and evaluation of biclustering methods for gene expression data. Bioinformatics 22, 1122–1129 (2006).
Sharan, R., Ulitsky, I. & Shamir, R. Network-based prediction of protein function. Mol. Syst. Biol. 3, 88 (2007).
Ideker, T., Ozier, O., Schwikowski, B. & Siegel, A. F. Discovering regulatory and signalling circuits in molecular interaction networks. Bioinformatics 18 (Suppl. 1), S233–S240 (2002).
Sohler, F., Hanisch, D. & Zimmer, R. New methods for joint analysis of biological networks and expression data. Bioinformatics 20, 1517–1521 (2004).
Cabusora, L., Sutton, E., Fulmer, A. & Forst, C. V. Differential network expression during drug and stress response. Bioinformatics 21, 2898–2905 (2005).
Scott, J., Ideker, T., Karp, R. M. & Sharan, R. Efficient algorithms for detecting signaling pathways in protein interaction networks. J. Comput. Biol. 13, 133–144 (2006).
Nacu, S., Critchley-Thorne, R., Lee, P. & Holmes, S. Gene expression network analysis and applications to immunology. Bioinformatics 23, 850–858 (2007).
Huang, S. S. & Fraenkel, E. Integrating proteomic, transcriptional, and interactome data reveals hidden components of signaling and regulatory networks. Sci. Signal. 2, ra40 (2009).
Chowdhury, S. A. & Koyutürk, M. Identification of coordinately dysregulated subnetworks in complex phenotypes. Pac. Symp. Biocomput. 2010, 133–144 (2010).
Dao, P. et al. Optimally discriminative subnetwork markers predict response to chemotherapy. Bioinformatics 27, i205–i213 (2011).
Fortney, K., Kotlyar, M. & Jurisica, I. Inferring the functions of longevity genes with modular subnetwork biomarkers of Caenorhabditis elegans aging. Genome Biol. 11, R13 (2010).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003).
Ulitsky, I. & Shamir, R. Identifying functional modules using expression profiles and confidence-scored protein interactions. Bioinformatics 25, 1158–1164 (2009).
Wang, Y. C. & Chen, B. S. Integrated cellular network of transcription regulations and protein-protein interactions. BMC Syst. Biol. 4, 20 (2010).
Breitling, R., Amtmann, A. & Herzyk, P. Graph-based iterative Group Analysis enhances microarray interpretation. BMC Bioinformatics 5, 100 (2004).
Rajagopalan, D. & Agarwal, P. Inferring pathways from gene lists using a literature-derived network of biological relationships. Bioinformatics 21, 788–793 (2005).
Chuang, H. Y., Lee, E., Liu, Y. T., Lee, D. & Ideker, T. Network-based classification of breast cancer metastasis. Mol. Syst. Biol. 3, 140 (2007).
Hwang, T. & Park, T. Identification of differentially expressed subnetworks based on multivariate ANOVA. BMC Bioinformatics 10, 128 (2009).
Klammer, M., Godl, K., Tebbe, A. & Schaab, C. Identifying differentially regulated subnetworks from phosphoproteomic data. BMC Bioinformatics 11, 351 (2010).
Zhao, X. M., Wang, R. S., Chen, L. & Aihara, K. Uncovering signal transduction networks from high-throughput data by integer linear programming. Nucleic Acids Res. 36, e48 (2008).
Backes, C. et al. An integer linear programming approach for finding deregulated subgraphs in regulatory networks. Nucleic Acids Res. 40, e43 (2012).
Vandin, F., Upfal, E. & Raphael, B. J. Algorithms for detecting significantly mutated pathways in cancer. J. Comput. Biol. 18, 507–522 (2011).
Komurov, K., White, M. A. & Ram, P. T. Use of data-biased random walks on graphs for the retrieval of context-specific networks from genomic data. PLoS Comput. Biol. 6, e1000889 (2010).
Reiss, D. J., Baliga, N. S. & Bonneau, R. Integrated biclustering of heterogeneous genome-wide datasets for the inference of global regulatory networks. BMC Bioinformatics 7, 280 (2006).
Vaske, C. J. et al. Inference of patient-specific pathway activities from multi-dimensional cancer genomics data using PARADIGM. Bioinformatics 26, i237–i245 (2010).
Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature 474, 609–615 (2011).
Cancer Genome Atlas Research Network. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 499, 43–49 (2013). A genome-scale effort that mapped significantly mutated pathways in human cancer through network projection of mutational profiles, leading to the identification of novel disease mechanisms.
Ciriello, G., Cerami, E., Sander, C. & Schultz, N. Mutual exclusivity analysis identifies oncogenic network modules. Genome Res. 22, 398–406 (2012).
Miller, C. A., Settle, S. H., Sulman, E. P., Aldape, K. D. & Milosavljevic, A. Discovering functional modules by identifying recurrent and mutually exclusive mutational patterns in tumors. BMC Med. Genom. 4, 34 (2011).
Lan, A. et al. ResponseNet: revealing signaling and regulatory networks linking genetic and transcriptomic screening data. Nucleic Acids Res. 39, W424–W429 (2011).
Komurov, K., Dursun, S., Erdin, S. & Ram, P. T. NetWalker: a contextual network analysis tool for functional genomics. BMC Genomics 13, 282 (2012).
Rives, A. W. & Galitski, T. Modular organization of cellular networks. Proc. Natl Acad. Sci. USA 100, 1128–1133 (2003).
Ravasz, E. & Barabasi, A. L. Hierarchical organization in complex networks. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 67, 026112 (2003).
Hanisch, D., Zien, A., Zimmer, R. & Lengauer, T. Co-clustering of biological networks and gene expression data. Bioinformatics 18 (Suppl. 1), S145–S154 (2002).
Gonzalez, O. & Zimmer, R. Contextual analysis of RNAi-based functional screens using interaction networks. Bioinformatics 27, 2707–2713 (2011).
Luscombe, N. M. et al. Genomic analysis of regulatory network dynamics reveals large topological changes. Nature 431, 308–312 (2004). This article presents an omics-based mapping of stress-response pathways in yeast protein networks, revealing key regulatory insights into network dynamics.
Tanay, A., Sharan, R., Kupiec, M. & Shamir, R. Revealing modularity and organization in the yeast molecular network by integrated analysis of highly heterogeneous genomewide data. Proc. Natl Acad. Sci. USA 101, 2981–2986 (2004). This study develops and integrates a widely cited approach for module discovery that allows the simultaneous interpretation of a diverse range of biological information.
Blazier, A. S. & Papin, J. A. Integration of expression data in genome-scale metabolic network reconstructions. Front. Physiol. 3, 299 (2012).
Lewis, N. E., Nagarajan, H. & Palsson, B. O. Constraining the metabolic genotype-phenotype relationship using a phylogeny of in silico methods. Nature Rev. Microbiol. 10, 291–305 (2012).
Schellenberger, J. et al. Quantitative prediction of cellular metabolism with constraint-based models: the COBRA Toolbox v2.0. Nature Protoc. 6, 1290–1307 (2011).
Calvano, S. E. et al. A network-based analysis of systemic inflammation in humans. Nature 437, 1032–1037 (2005).
Yeger-Lotem, E. et al. Bridging high-throughput genetic and transcriptional data reveals cellular responses to α−synuclein toxicity. Nature Genet. 41, 316–323 (2009).
Xue, H. et al. A modular network model of aging. Mol. Syst. Biol. 3, 147 (2007).
Bandyopadhyay, S., Kelley, R. & Ideker, T. Discovering regulated networks during HIV-1 latency and reactivation. Pac. Symp. Biocomput. 2006, 354–366 (2006).
Haugen, A. C. et al. Integrating phenotypic and expression profiles to map arsenic-response networks. Genome Biol. 5, R95 (2004).
Shlomi, T., Cabili, M. N., Herrgard, M. J., Palsson, B. O. & Ruppin, E. Network-based prediction of human tissue-specific metabolism. Nature Biotech. 26, 1003–1010 (2008).
Colijn, C. et al. Interpreting expression data with metabolic flux models: predicting Mycobacterium tuberculosis mycolic acid production. PLoS Comput. Biol. 5, e1000489 (2009).
Price, N. D., Reed, J. L. & Palsson, B. O. Genome-scale models of microbial cells: evaluating the consequences of constraints. Nature Rev. Microbiol. 2, 886–897 (2004).
Chowdhury, S. A., Nibbe, R. K., Chance, M. R. & Koyuturk, M. Subnetwork state functions define dysregulated subnetworks in cancer. J. Comput. Biol. 18, 263–281 (2011).
Anastassiou, D. Computational analysis of the synergy among multiple interacting genes. Mol. Syst. Biol. 3, 83 (2007).
Ma, X., Lee, H., Wang, L. & Sun, F. CGI: a new approach for prioritizing genes by combining gene expression and protein-protein interaction data. Bioinformatics 23, 215–221 (2007).
Li, W. et al. Dynamical systems for discovering protein complexes and functional modules from biological networks. IEEE/ACM Trans. Comput. Biol. Bioinform. 4, 233–250 (2007).
Yang, P., Li, X., Wu, M., Kwoh, C. K. & Ng, S. K. Inferring gene-phenotype associations via global protein complex network propagation. PLoS ONE 6, e21502 (2011).
Tu, Z. et al. Integrating siRNA and protein-protein interaction data to identify an expanded insulin signaling network. Genome Res. 19, 1057–1067 (2009).
Taylor, I. W. et al. Dynamic modularity in protein interaction networks predicts breast cancer outcome. Nature Biotech. 27, 199–204 (2009). An omics-based strategy for identifying breast cancer pathways, which demonstrated the power of integrative network analysis for disease prognosis.
Zhang, X. et al. The expanded human disease network combining protein-protein interaction information. Eur. J. Hum. Genet. 19, 783–788 (2011).
Bapat, S. A., Krishnan, A., Ghanate, A. D., Kusumbe, A. P. & Kalra, R. S. Gene expression: protein interaction systems network modeling identifies transformation-associated molecules and pathways in ovarian cancer. Cancer Res. 70, 4809–4819 (2010).
Zhang, K. X. & Ouellette, B. F. CAERUS: predicting CAncER oUtcomeS using relationship between protein structural information, protein networks, gene expression data, and mutation data. PLoS Comput. Biol. 7, e1001114 (2011).
Ma, H., Schadt, E. E., Kaplan, L. M. & Zhao, H. COSINE: COndition-SpecIfic sub-NEtwork identification using a global optimization method. Bioinformatics 27, 1290–1298 (2011).
Ahn, J., Yoon, Y., Park, C., Shin, E. & Park, S. Integrative gene network construction for predicting a set of complementary prostate cancer genes. Bioinformatics 27, 1846–1853 (2011).
Wu, Z., Zhao, X. M. & Chen, L. A systems biology approach to identify effective cocktail drugs. BMC Syst. Biol. 4, (Suppl. 2), S7 (2010).
Vespignani, A. Evolution thinks modular. Nature Genet. 35, 118–119 (2003).
Mazurie, A., Bonchev, D., Schwikowski, B. & Buck, G. A. Evolution of metabolic network organization. BMC Syst. Biol. 4, 59 (2010).
Odom, D. T. et al. Tissue-specific transcriptional regulation has diverged significantly between human and mouse. Nature Genet. 39, 730–732 (2007).
Wuchty, S., Oltvai, Z. N. & Barabasi, A. L. Evolutionary conservation of motif constituents in the yeast protein interaction network. Nature Genet. 35, 176–179 (2003).
Matthews, L. R. et al. Identification of potential interaction networks using sequence-based searches for conserved protein-protein interactions or “interologs”. Genome Res. 11, 2120–2126 (2001).
Yu, H. et al. Annotation transfer between genomes: protein-protein interologs and protein-DNA regulogs. Genome Res. 14, 1107–1118 (2004).
Sharan, R. & Ideker, T. Modeling cellular machinery through biological network comparison. Nature Biotech. 24, 427–433 (2006).
Kelley, B. P. et al. PathBLAST: a tool for alignment of protein interaction networks. Nucleic Acids Res. 32, W83–W88 (2004).
Sharan, R. et al. Conserved patterns of protein interaction in multiple species. Proc. Natl Acad. Sci. USA 102, 1974–1979 (2005). This study highlights a method for the pairwise alignment of subnetworks to facilitate efficient comparisons between diverse interactomes.
Kalaev, M., Bafna, V. & Sharan, R. Fast and accurate alignment of multiple protein networks. J. Comput. Biol. 16, 989–999 (2009).
Bandyopadhyay, S., Sharan, R. & Ideker, T. Systematic identification of functional orthologs based on protein network comparison. Genome Res. 16, 428–435 (2006).
Singh, R., Xu, J. & Berger, B. Global alignment of multiple protein interaction networks with application to functional orthology detection. Proc. Natl Acad. Sci. USA 105, 12763–12768 (2008).
Flannick, J., Novak, A., Srinivasan, B. S., McAdams, H. H. & Batzoglou, S. Graemlin: general and robust alignment of multiple large interaction networks. Genome Res. 16, 1169–1181 (2006).
Berg, J. & Lassig, M. Cross-species analysis of biological networks by Bayesian alignment. Proc. Natl Acad. Sci. USA 103, 10967–10972 (2006).
Barabasi, A. L. & Albert, R. Emergence of scaling in random networks. Science 286, 509–512 (1999).
Arabidopsis Interactome Mapping Consortium. Evidence for network evolution in an Arabidopsis interactome map. Science 333, 601–607 (2011).
Koyuturk, M. et al. Pairwise alignment of protein interaction networks. J. Comput. Biol. 13, 182–199 (2006).
Dutkowski, J. & Tiuryn, J. Identification of functional modules from conserved ancestral protein-protein interactions. Bioinformatics 23, i149–i158 (2007).
Raymond, J. & Segre, D. The effect of oxygen on biochemical networks and the evolution of complex life. Science 311, 1764–1767 (2006).
Vazquez, A., Flammini, A., Maritan, A. & Vespignani, A. Global protein function prediction from protein-protein interaction networks. Nature Biotech. 21, 697–700 (2003).
Sharan, R., Ideker, T., Kelley, B., Shamir, R. & Karp, R. M. Identification of protein complexes by comparative analysis of yeast and bacterial protein interaction data. J. Comput. Biol. 12, 835–846 (2005).
Ulitsky, I. & Shamir, R. Pathway redundancy and protein essentiality revealed in the Saccharomyces cerevisiae interaction networks. Mol. Syst. Biol. 3, 104 (2007).
Ideker, T. & Krogan, N. J. Differential network biology. Mol. Syst. Biol. 8, 565 (2012).
Kapitzky, L. et al. Cross-species chemogenomic profiling reveals evolutionarily conserved drug mode of action. Mol. Syst. Biol. 6, 451 (2010).
Suthram, S., Sittler, T. & Ideker, T. The Plasmodium protein network diverges from those of other eukaryotes. Nature 438, 108–112 (2005).
Hillenmeyer, M. E. et al. The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science 320, 362–365 (2008).
Han, J. D. et al. Evidence for dynamically organized modularity in the yeast protein-protein interaction network. Nature 430, 88–93 (2004).
Andreopoulos, B., An, A., Wang, X. & Schroeder, M. A roadmap of clustering algorithms: finding a match for a biomedical application. Brief Bioinform. 10, 297–314 (2009).
Saito, R. et al. A travel guide to Cytoscape plugins. Nature Methods 9, 1069–1076 (2012).
Yosef, N. et al. ANAT: a tool for constructing and analyzing functional protein networks. Sci. Signal. 4, pl1 (2011).
Workman, C. T. et al. A systems approach to mapping DNA damage response pathways. Science 312, 1054–1059 (2006).
Bisson, N. et al. Selected reaction monitoring mass spectrometry reveals the dynamics of signaling through the GRB2 adaptor. Nature Biotech. 29, 653–658 (2011).
Ellis, J. D. et al. Tissue-specific alternative splicing remodels protein-protein interaction networks. Mol. Cell 46, 884–892 (2012).
Guenole, A. et al. Dissection of DNA damage responses using multiconditional genetic interaction maps. Mol. Cell 49, 346–358 (2013).
Altay, G., Asim, M., Markowetz, F. & Neal, D. E. Differential C3NET reveals disease networks of direct physical interactions. BMC Bioinformatics 12, 296 (2011).
Valcarcel, B. et al. A differential network approach to exploring differences between biological states: an application to prediabetes. PLoS ONE 6, e24702 (2011).
Beyer, A., Bandyopadhyay, S. & Ideker, T. Integrating physical and genetic maps: from genomes to interaction networks. Nature Rev. Genet. 8, 699–710 (2007).
Kelley, R. & Ideker, T. Systematic interpretation of genetic interactions using protein networks. Nature Biotech. 23, 561–566 (2005).
Ulitsky, I., Shlomi, T., Kupiec, M. & Shamir, R. From E-MAPs to module maps: dissecting quantitative genetic interactions using physical interactions. Mol. Syst. Biol. 4, 209 (2008).
Bandyopadhyay, S., Kelley, R., Krogan, N. J. & Ideker, T. Functional maps of protein complexes from quantitative genetic interaction data. PLoS Comput. Biol. 4, e1000065 (2008).
Srivas, R. et al. Assembling global maps of cellular function through integrative analysis of physical and genetic networks. Nature Protoc. 6, 1308–1323 (2011).
Zhao, R. et al. Navigating the chaperone network: an integrative map of physical and genetic interactions mediated by the Hsp90 chaperone. Cell 120, 715–727 (2005).
Wilmes, G. M. et al. A genetic interaction map of RNA-processing factors reveals links between Sem1/Dss1-containing complexes and mRNA export and splicing. Mol. Cell 32, 735–746 (2008).
Fiedler, D. et al. Functional organization of the S. cerevisiae phosphorylation network. Cell 136, 952–963 (2009).
Zhang, L. V. et al. Motifs, themes and thematic maps of an integrated Saccharomyces cerevisiae interaction network. J. Biol. 4, 6 (2005).
Yeger-Lotem, E. et al. Network motifs in integrated cellular networks of transcription-regulation and protein-protein interaction. Proc. Natl Acad. Sci. USA 101, 5934–5939 (2004).
Tan, K., Shlomi, T., Feizi, H., Ideker, T. & Sharan, R. Transcriptional regulation of protein complexes within and across species. Proc. Natl Acad. Sci. USA 104, 1283–1288 (2007).
Herrgard, M. J., Lee, B. S., Portnoy, V. & Palsson, B. O. Integrated analysis of regulatory and metabolic networks reveals novel regulatory mechanisms in Saccharomyces cerevisiae. Genome Res. 16, 627–635 (2006).
Lee, J. M., Gianchandani, E. P., Eddy, J. A. & Papin, J. A. Dynamic analysis of integrated signaling, metabolic, and regulatory networks. PLoS Comput. Biol. 4, e1000086 (2008).
Chandrasekaran, S. & Price, N. D. Probabilistic integrative modeling of genome-scale metabolic and regulatory networks in Escherichia coli and Mycobacterium tuberculosis. Proc. Natl Acad. Sci. USA 107, 17845–17850 (2010).
Deshpande, R., Sharma, S., Verfaillie, C. M., Hu, W. S. & Myers, C. L. A scalable approach for discovering conserved active subnetworks across species. PLoS Comput. Biol. 6, e1001028 (2010). This study illustrates how a combination of integrative approaches may be simultaneously applied for the identification of conserved active modules.
Waltman, P. et al. Multi-species integrative biclustering. Genome Biol. 11, R96 (2010).
Ryan, C. J. et al. Hierarchical modularity and the evolution of genetic interactomes across species. Mol. Cell 46, 691–704 (2012).
Stark, C. et al. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 34, D535–D539 (2006).
Ito, T. et al. Roles for the two-hybrid system in exploration of the yeast protein interactome. Mol. Cell. Proteomics. 1, 561–566 (2002).
Shou, C. et al. Measuring the evolutionary rewiring of biological networks. PLoS Comput. Biol. 7, e1001050 (2011).
Lee, I., Blom, U. M., Wang, P. I., Shim, J. E. & Marcotte, E. M. Prioritizing candidate disease genes by network-based boosting of genome-wide association data. Genome Res. 21, 1109–1121 (2011).
Bebek, G., Koyuturk, M., Price, N. D. & Chance, M. R. Network biology methods integrating biological data for translational science. Brief Bioinform. 13, 446–459 (2012).
Ulitsky, I. & Shamir, R. Identification of functional modules using network topology and high-throughput data. BMC Syst. Biol. 1, 8 (2007).
Beisser, D., Klau, G. W., Dandekar, T., Müller, T. & Dittrich, M. T. BioNet: an R-Package for the functional analysis of biological networks. Bioinformatics 26, 1129–1130 (2010).
Paull, E. O. et al. Discovering causal pathways linking genomic events to transcriptional states using Tied Diffusion Through Interacting Events (TieDIE). Bioinformatics http://dx.doi.org/10.1093/bioinformatics/btt471 (2013).
Kalaev, M., Smoot, M., Ideker, T. & Sharan, R. NetworkBLAST: comparative analysis of protein networks. Bioinformatics 24, 594–596 (2008).
Liao, C. S., Lu, K., Baym, M., Singh, R. & Berger, B. IsoRankN: spectral methods for global alignment of multiple protein networks. Bioinformatics 25, i253–i258 (2009).
Zhang. B. et al. DDN: a caBIG® analytical tool for differential network analysis. Bioinformatics 27, 1036–1038 (2011).
Gill, R. & Datta, S. A statistical framework for differential network analysis from microarray data. BMC Bioinformatics 11, 95 (2010).
Acknowledgements
We gratefully acknowledge US National Institutes of Health (NIH) grants P41 GM103504 and P50 GM085764 in support of this work.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
FURTHER INFORMATION
Supplementary information
Supplementary information S1 (table)
Sources of molecular interaction and 'omics' profiling data (PDF 296 kb)
Glossary
- Epistasis
-
The phenomenon whereby the function of one gene affects the phenotype (for example, growth) of another gene in a non-additive manner.
- Synthetic lethality
-
An extreme case of negative genetic epistasis in which the mutation of two genes in combination, but not individually, causes a lethal phenotype.
- Degree
-
The number of interactions (edges) that a molecule (node) has in a network.
- Betweenness centrality
-
A statistical intuition of how 'central' the status of a given molecule (node) or interaction (edge) is within a network. This is inferred by the fraction of shortest paths between all pairs of nodes that pass through a particular node or edge.
- Network topology
-
The overall arrangement of nodes and edges in a given network.
- Metabolic flux
-
The flow of chemicals through any metabolic reaction (for example, an enzymatic reaction).
- Hubs
-
Molecules with the highest number of interactions (degree) in a network.
- Orthologous
-
Refers to the evolutionary relationship between two genes in two species that have descended from a common ancestor. Such genes are denoted as orthologues.
Rights and permissions
About this article
Cite this article
Mitra, K., Carvunis, AR., Ramesh, S. et al. Integrative approaches for finding modular structure in biological networks. Nat Rev Genet 14, 719–732 (2013). https://doi.org/10.1038/nrg3552
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrg3552
This article is cited by
-
A novel approach for predicting upstream regulators (PURE) that affect gene expression
Scientific Reports (2023)
-
Omics correlation for efficient network construction
Nature Computational Science (2023)
-
Mena regulates nesprin-2 to control actin–nuclear lamina associations, trans-nuclear membrane signalling and gene expression
Nature Communications (2023)
-
Protein interaction networks provide insight into fetal origins of chronic obstructive pulmonary disease
Respiratory Research (2022)
-
Mendelian randomization of circulating proteome identifies actionable targets in heart failure
BMC Genomics (2022)