Key Points
-
This article focuses on the aims, methodological requisites and study designs of cancer genome-sequencing studies. Thus far, most cancer genome-sequencing studies have had the aims of discovering driver mutations, identifying somatic mutational signatures, characterizing clonal evolution and/or advancing personalized medicine.
-
Key aspects of second-generation cancer genome-sequencing study design include full sequencing of the matched normal genome of the cancer type, at least 30-fold redundant sequence coverage for the detection of inherited and somatic single-nucleotide variation and verification resequencing to confirm the somatic status of acquired mutations.
-
Single-patient studies are hypothesis-generating and have the potential to inform clinical practice, but they do not allow for generalization of findings. The discovery cohort is a group of cancers of the same type, or subtype, subject to second-generation sequencing. Discovery cohort studies have the potential to detect recurrent somatic mutations of genes and pathways.
-
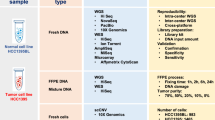
Multi-ome discovery cohorts use second-generation technology to sequence an assortment of genomes, exomes and/or transcriptomes in a group of cancers of the same type or subtype.
Abstract
Discoveries from cancer genome sequencing have the potential to translate into advances in cancer prevention, diagnostics, prognostics, treatment and basic biology. Given the diversity of downstream applications, cancer genome-sequencing studies need to be designed to best fulfil specific aims. Knowledge of second-generation cancer genome-sequencing study design also facilitates assessment of the validity and importance of the rapidly growing number of published studies. In this Review, we focus on the practical application of second-generation sequencing technology (also known as next-generation sequencing) to cancer genomics and discuss how aspects of study design and methodological considerations — such as the size and composition of the discovery cohort — can be tailored to serve specific research aims.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Stratton, M. R., Campbell, P. J. & Futreal, P. A. The cancer genome. Nature 458, 719–724 (2009).
Ley, T. J. et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature 456, 66–72 (2008). This was first study to use second-generation technology to sequence a cancer genome. It established cancer genome sequencing as an unbiased method for discovering candidate driver mutations.
International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature 431, 931–945 (2004).
Lander, E. S. et al. Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001).
Battelle Technology Partnership Practice. Economic impact of the Human Genome Project: how a $3.8 billion investment drove $796 billion in economic impact, created 310,000 jobs, and launched the genomic revolution. battelle.org[online], (2011).
Morin, R. D. et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nature Genet. 42, 181–185 (2010) (2011).
Sneeringer, C. J. et al. Coordinated activities of wild-type plus mutant EZH2 drive tumor-associated hypertrimethylation of lysine 27 on histone H3 (H3K27) in human B-cell lymphomas. Proc. Natl Acad. Sci. USA 107, 20980–20985 (2010).
McCabe, M. T. et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature 492, 108–112 (2012).
Nik-Zainal, S. et al. Mutational processes molding the genomes of 21 breast cancers. Cell 149, 979–993 (2012).
Stephens, P. J. et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell 144, 27–40 (2011).
Misale, S. et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature 486, 532–536 (2012).
Northcott, P. A. et al. Medulloblastomics: the end of the beginning. Nature Rev. Cancer 12, 818–834 (2012).
Meyerson, M., Gabriel, S. & Getz, G. Advances in understanding cancer genomes through second-generation sequencing. Nature Rev. Genet. 11, 685–696 (2010).
Mardis, E. R. et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N. Engl. J. Med. 361, 1058–1066 (2009).
Link, D. C. Identification of a novel TP53 cancer susceptibility mutation through whole-genome sequencing of a patient with therapy-related AML. JAMA 305, 1568 (2011).
Shah, S. P. et al. Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature 461, 809–813 (2009). This study used ultra-deep resequencing to characterize clonal evolution and showed that variable somatic mutation allele frequencies can reflect different subclones. Moreover, considerable evolution can occur over time.
Jones, S. J. et al. Evolution of an adenocarcinoma in response to selection by targeted kinase inhibitors. Genome Biol. 11, R82 (2010). This work incorporated second-generation sequencing into the personalized medicine framework. Specifically, the intent of the case study was to inform physician decision making with respect to treatment of a rare cancer.
Ding, L. et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature 464, 999–1005 (2010).
Pleasance, E. D. et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature 463, 191–196 (2009).
Pleasance, E. D. et al. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature 463, 184–190 (2009). This study highlighted that the distribution and composition of somatic mutations across a genome is not uniform. It showed that through examining the mutational signatures, researchers can gain insight into the mechanisms and processes that may have given rise to the mutations.
Lee, W. et al. The mutation spectrum revealed by paired genome sequences from a lung cancer patient. Nature 465, 473–477 (2010).
Levy, S. et al. The diploid genome sequence of an individual human. PLoS Biol. 5, e254 (2007).
Wheeler, D. A. et al. The complete genome of an individual by massively parallel DNA sequencing. Nature 452, 872–876 (2008).
Wang, J. et al. The diploid genome sequence of an Asian individual. Nature 456, 60–65 (2008).
Bentley, D. R. et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature 456, 53–59 (2008). An accurate consensus sequence was built with second-generation technology from >30-fold redundant coverage of 35 bp paired-end reads.
Pelak, K. et al. The characterization of twenty sequenced human genomes. PLoS Genet. 6, e1001111 (2010).
Ellis, M. J. et al. Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature 486, 353–360 (2012).
Bass, A. J. et al. Genomic sequencing of colorectal adenocarcinomas identifies a recurrent VTI1A-TCF7L2 fusion. Nature Genet. 43, 964–968 (2011).
Berger, M. F. et al. The genomic complexity of primary human prostate cancer. Nature 470, 214–220 (2011).
Fujimoto, A. et al. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nature Genet. 44, 760–764 (2012).
Sherry, S. T. et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 29, 308–311 (2001).
Ajay, S. S., Parker, S. C. J., Ozel Abaan, H., Fuentes Fajardo, K. V. & Margulies, E. H. Accurate and comprehensive sequencing of personal genomes. Genome Res. 21, 1498–1505 (2011).
Navin, N. et al. Tumour evolution inferred by single-cell sequencing. Nature 472, 90–94 (2011).
Turajlic, S. et al. Whole genome sequencing of matched primary and metastatic acral melanomas. Genome Res. 22, 196–207 (2011).
Puente, X. S. et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature 475, 101–105 (2011).
Campbell, P. J. et al. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature 467, 1109–1113 (2010).
Peña-Llopis, S. et al. BAP1 loss defines a new class of renal cell carcinoma. Nature Genet. 44, 751–759 (2012).
Ng, C. K. et al. The role of tandem duplicator phenotype in tumour evolution in high-grade serous ovarian cancer. J. Pathol. 226, 703–712 (2012).
Stephens, P. J. et al. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature 462, 1005–1010 (2009).
Kloosterman, W. P. et al. Chromothripsis is a common mechanism driving genomic rearrangements in primary and metastatic colorectal cancer. Genome Biol. 12, R103 (2011).
McBride, D. J. et al. Tandem duplication of chromosomal segments is common in ovarian and breast cancer genomes. J. Pathol. 227, 446–455 (2012).
Campbell, P. J. et al. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nature Genet. 40, 722–729 (2008). By investigating the paired-end sequencing reads that did not align to the reference genome as expected with respect to each other, the authors were able to demonstrate a high-throughput and high-resolution bioinformatics method to characterize structural variation.
Muzny, D. M. et al. Comprehensive molecular characterization of human colon and rectal cancer. Nature 487, 330–337 (2012). With 97 colorectal cancer genomes sequenced to low-to-moderate redundant coverage, this discovery cohort is the largest to date.
Korbel, J. O. et al. Paired-end mapping reveals extensive structural variation in the human genome. Science 318, 420–426 (2007).
Onishi-Seebacher, M. & Korbel, J. O. Challenges in studying genomic structural variant formation mechanisms: the short-read dilemma and beyond. BioEssays 33, 840–850 (2011).
Simpson, J. T. et al. ABySS: a parallel assembler for short read sequence data. Genome Res. 19, 1117–1123 (2009).
Fullwood, M. J., Wei, C.-L., Liu, E. T. & Ruan, Y. Next-generation DNA sequencing of paired-end tags (PET) for transcriptome and genome analyses. Genome Res. 19, 521–532 (2009).
Medvedev, P., Stanciu, M. & Brudno, M. Computational methods for discovering structural variation with next-generation sequencing. Nature Methods 6, S13–S20 (2009).
Hillmer, A. M. et al. Comprehensive long-span paired-end-tag mapping reveals characteristic patterns of structural variations in epithelial cancer genomes. Genome Res. 21, 665–675 (2011).
Boeva, V. et al. Control-FREEC: a tool for assessing copy number and allelic content using next-generation sequencing data. Bioinformatics 28, 423–425 (2012).
Druker, B. J. et al. Five-year follow-up of patients receiving imatinib for chronic myeloid leukemia. N. Engl. J. Med. 355, 2408–2417 (2006).
Lee, E. et al. Landscape of somatic retrotransposition in human cancers. Science 337, 967–971 (2012).
Welch, J. S. Use of whole-genome sequencing to diagnose a cryptic fusion oncogene. JAMA 305, 1577 (2011).
Weiss, G. J. et al. Paired tumor and normal whole genome sequencing of metastatic olfactory neuroblastoma. PLoS ONE 7, e37029 (2012).
Tao, Y. et al. Rapid growth of a hepatocellular carcinoma and the driving mutations revealed by cell-population genetic analysis of whole-genome data. Proc. Natl Acad. Sci. USA 108, 12042–12047 (2011).
Bueno, R. et al. Second generation sequencing of the mesothelioma tumor genome. PLoS ONE 5, e10612 (2010).
Totoki, Y. et al. High-resolution characterization of a hepatocellular carcinoma genome. Nature Genet. 43, 464–469 (2011).
Demeure, M. J. et al. Cancer of the ampulla of Vater: analysis of the whole genome sequence exposes a potential therapeutic vulnerability. Genome Med. 4, 56 (2012).
Muller, F. L. et al. Passenger deletions generate therapeutic vulnerabilities in cancer. Nature 488, 337–342 (2012).
Shah, S. P. et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature 486, 395–399 (2012).
Nik-Zainal, S. et al. The life history of 21 breast cancers. Cell 149, 994–1007 (2012). This paper demonstrates the utility of characterizing the somatic mutational signature with the discovery of kataegis.
Morin, R. D. et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature 476, 298–303 (2011).
Wu, G. et al. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nature Genet. 44, 251–253 (2012).
Wang, L. et al. SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. N. Engl. J. Med. 365, 2497–2506 (2011).
Chapman, M. A. et al. Initial genome sequencing and analysis of multiple myeloma. Nature 471, 467–472 (2011).
Harbour, J. W. et al. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science 330, 1410–1413 (2010). This study discovered a gene that was somatically mutated in an impressive number of metastasizing tumours using second-generation sequencing of exomes. This study highlights that there are novel and valuable candidate therapeutic targets that are yet to be discovered.
Yoshida, K. et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature 478, 64–69 (2011).
Schwartzentruber, J. et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 482, 226–231 (2012).
Pugh, T. J. et al. Medulloblastoma exome sequencing uncovers subtype-specific somatic mutations. Nature 488, 106–110 (2012).
Sathirapongsasuti, J. F. et al. Exome sequencing-based copy-number variation and loss of heterozygosity detection: ExomeCNV. Bioinformatics 27, 2648–2654 (2011).
Karakoc, E. et al. Detection of structural variants and indels within exome data. Nature Methods 9, 176–178 (2012).
Banerji, S. et al. Sequence analysis of mutations and translocations across breast cancer subtypes. Nature 486, 405–409 (2012).
Ruan, Y. et al. Fusion transcripts and transcribed retrotransposed loci discovered through comprehensive transcriptome analysis using paired-end ditags (PETs). Genome Res. 17, 828–838 (2007).
Mortazavi, A., Williams, B. A., McCue, K., Schaeffer, L. & Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-seq. Nature Methods 5, 621–628 (2008).
Roberts, K. G. et al. Genetic alterations activating kinase and cytokine receptor signaling in high-risk acute lymphoblastic leukemia. Cancer Cell 22, 153–166 (2012).
Jones, D. T. W. et al. Dissecting the genomic complexity underlying medulloblastoma. Nature 488, 100–105 (2012).
Hammerman, P. S. et al. Comprehensive genomic characterization of squamous cell lung cancers. Nature 489, 519–525 (2012).
Morin, R. D. et al. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 18, 610–621 (2008).
Wang, Z., Gerstein, M. & Snyder, M. RNA-seq: a revolutionary tool for transcriptomics. Nature Rev. Genet. 10, 57–63 (2009).
Dees, N. D. et al. MuSiC: identifying mutational significance in cancer genomes. Genome Res. 22, 1589–1598 (2012).
Vaske, C. J. et al. Inference of patient-specific pathway activities from multi-dimensional cancer genomics data using PARADIGM. Bioinformatics 26, i237–i245 (2010).
Hawkins, R. D., Hon, G. C. & Ren, B. Next-generation genomics: an integrative approach. Nature Rev. Genet. 11, 476–486 (2010).
Zhang, J. et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature 481, 157–163 (2012).
Ju, Y. S. et al. A transforming KIF5B and RET gene fusion in lung adenocarcinoma revealed from whole-genome and transcriptome sequencing. Genome Res. 22, 436–445 (2011).
Esteller, M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nature Rev. Genet. 8, 286–298 (2007).
Barski, A. et al. High-resolution profiling of histone methylations in the human genome. Cell 129, 823–837 (2007).
Lister, R. et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462, 315–322 (2009).
Zhang, J. et al. A novel retinoblastoma therapy from genomic and epigenetic analyses. Nature 481, 329–334 (2012).
Cheung, N.-K. V. et al. Association of age at diagnosis and genetic mutations in patients with neuroblastoma. JAMA 307, 1062–1071 (2012).
Molenaar, J. J. et al. Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature 483, 589–593 (2012). This is one of the largest discovery cohorts to date. Researchers sequenced the genomes of 87 tumour–normal pairs to at least 30-fold redundant coverage.
Collins, C. C. et al. Next generation sequencing of prostate cancer from a patient identifies a deficiency of methylthioadenosine phosphorylase, an exploitable tumor target. Mol. Cancer Ther. 11, 775–783 (2012).
Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011).
Rausch, T. et al. Genome sequencing of pediatric medulloblastoma links catastrophic DNA rearrangements with TP53 mutations. Cell 148, 59–71 (2012).
Sung, W.-K. et al. Genome-wide survey of recurrent HBV integration in hepatocellular carcinoma. Nature Genet. 44, 765–769 (2012).
Klein, G. Lymphoma development in mice and humans: diversity of initiation is followed by convergent cytogenetic evolution. Proc. Natl Acad. Sci. USA 76, 2442–2446 (1979).
Castoe, T. A., De Koning, A. P. J. & Pollock, D. D. Adaptive molecular convergence: molecular evolution versus molecular phylogenetics. Commun. Integr. Biol. 3, 67–69 (2010).
Berger, M. F. et al. Melanoma genome sequencing reveals frequent PREX2 mutations. Nature 485, 502–506 (2012).
Kimchi-Sarfaty, C. et al. A 'silent' polymorphism in the MDR1 gene changes substrate specificity. Science 315, 525–528 (2007).
Pagani, F., Raponi, M. & Baralle, F. E. Synonymous mutations in CFTR exon 12 affect splicing and are not neutral in evolution. Proc. Natl Acad. Sci. USA 102, 6368–6372 (2005).
Ding, L. et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole- genome sequencing. Nature 481, 506–510 (2012).
Wu, C. et al. Integrated genome and transcriptome sequencing identifies a novel form of hybrid and aggressive prostate cancer. J. Pathol. 227, 53–61 (2012).
Gerlinger, M. et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 366, 883–892 (2012).
Walter, M. J. et al. Clonal architecture of secondary acute myeloid leukemia. N. Engl. J. Med. 366, 1090–1098 (2012).
Jones, S. et al. Comparative lesion sequencing provides insights into tumor evolution. Proc. Natl Acad. Sci. USA 105, 4283–4288 (2008).
Greenman, C., Wooster, R., Futreal, P. A., Stratton, M. R. & Easton, D. F. Statistical analysis of pathogenicity of somatic mutations in cancer. Genetics 173, 2187–2198 (2006).
Acknowledgements
J.C.M. thanks the Canadian Institutes of Health Research and the Michael Smith Foundation for Health Research for their support. M.A.M. is the University of British Columbia, Canada Research Chair in Genome Science.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
FURTHER INFORMATION
Supplementary information
Supplementary informtation S1 (table)
Second-generation sequencing cancer genomics study aims (PDF 155 kb)
Supplementary informtation S2 (table)
Second-generation sequencing cancer genomics methodological requisites (PDF 115 kb)
Supplementary informtation S3 (table)
Second-generation sequencing cancer genomics study design (PDF 141 kb)
Glossary
- Driver mutations
-
Somatic mutations that have a role in creating, controlling and/or directing some aspect of the cancer phenotype.
- Kataegis
-
From the Greek meaning 'thunderstorm', this refers to clusters of somatic single-nucleotide variants that often colocalize with somatic structural variants.
- Chromothripsis
-
From the Greek meaning 'chromosome shattering', this refers to a single event of genome shattering and reassembly that results in complex somatic structural variations characterized by oscillating copy number and tens to hundreds of rearrangements that localize to one or a few chromosomes.
- Redundant sequence coverage
-
The total number of bases sequenced divided by the total number of bases in the haploid genome.
- B allele frequencies
-
Frequencies equal to B/(A+B), where A is the count for the reference nucleotide at an inherited single-nucleotide polymorphism (SNP) position, and B is the count for the alternate nucleotide at that same SNP position.
- Paired-end reads
-
Sequencing reads from each end of the same DNA molecule. Knowing the sequence of both reads and the length of the DNA molecule improves mapping to a reference sequence, de novo assembly and detecting structural variations.
- Chimeric genes
-
A combination of segments of two or more genes that forms a new gene.
- Split reads
-
Sequencing reads that align to non-contiguous spans of the reference sequence owing to somatic structural variation.
- Mass-spectrometric genotyping
-
A method that generates locus-specific amplicons followed by primer extension that incorporates mass-modified dideoxynucleotides at the single-nucleotide polymorphism position. A mass spectrometer then measures the differential mass of the products.
- Multi-ome discovery cohort
-
A cohort of cancer genomes, exomes and/or transcriptomes; more than one omic measurement per sample is not necessary.
- Allelic imbalances
-
Unequal transcript levels of the alleles of a gene.
- Integration omics
-
Examining how somatic mutation or deregulation of a genome, transcriptome and/or epigenome converge on a pathway, process or gene; more than one omic measurement per sample is not necessary. For example, gene inactivation through single-nucleotide variants or epigenomic silencing.
- Interaction omics
-
Examining how the somatic mutation or deregulation of the genome, transcriptome and/or epigenome affect one another; more than one omic measurement per sample is ideal. For example, somatic copy number variants can have effects on transcript levels.
- Custom capture
-
Hybridization or amplification of selected regions of the genome to specifically capture loci for second-generation sequencing.
- Ultra-deep second-generation resequencing
-
Greater than 100-fold redundant sequence coverage of a targeted selection of somatic mutations.
- Multi-region sequencing
-
Sequencing of distinct regions of the same solid tumour, this allows for the examination of intra-tumour heterogeneity and clonal evolution.
- Clinically actionable drug targets
-
Biological molecules or processes that can be targeted by an existing or experimental drug.
Rights and permissions
About this article
Cite this article
Mwenifumbo, J., Marra, M. Cancer genome-sequencing study design. Nat Rev Genet 14, 321–332 (2013). https://doi.org/10.1038/nrg3445
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrg3445
This article is cited by
-
Clinical application of tumour-in-normal contamination assessment from whole genome sequencing
Nature Communications (2024)
-
MYEOV with High Frequencies of Mutations in Head and Neck Cancers Facilitates Cancer Cell Malignant Behaviors
Biochemical Genetics (2023)
-
Progress and challenges of sequencing and analyzing circulating tumor cells
Cell Biology and Toxicology (2018)
-
Representing genetic variation with synthetic DNA standards
Nature Methods (2016)
-
Leveraging a Multi-Omics Strategy for Prioritizing Personalized Candidate Mutation-Driver Genes: A Proof-of-Concept Study
Scientific Reports (2015)