Key Points
-
Synthetic gene circuits are designed to implement novel biologic function, including cellular logic, dynamics and complex cellular and multicellular behaviours. A decade after emerging as a discipline, synthetic biology is entering the mainstream of biological research in molecular and systems biology, biotechnology and biomedicine.
-
One important current frontier in synthetic biology is the design and implementation of circuits and networks which are larger and more sophisticated than those of the early years of the field. A formalized design process, which has been essential in other engineering disciplines for scaling to larger systems, is being developed.
-
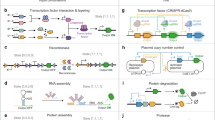
Design in synthetic biology combines top-down decomposition to break down complex problems into smaller subproblems with known solutions and bottom-up assembly, which combines components such as promoters, genes or higher-order modules into systems that solve the high-level problem.
-
At the level of molecular parts such as transcription factors, sensors and actuators, formalized design requires availability of large and compatible classes of components. Zinc finger and transcription-activator-like effector proteins, synthetic microRNAs and engineered cell surface receptors will each advance the field.
-
At the level of modules, much early work in synthetic biology has created and evaluated basic dynamic network motifs such as switches, oscillators and cascades, and an empirically informed choice of optimal circuit topologies for a given purposes is now possible. Cell–cell communication modules based on quorum sensing have been widely used; establishing similarly versatile modules in eukaryotic cells is now a priority. Eukaryotic signal processing based on protein–protein interactions has also been engineered.
-
A small number of large, sophisticated, integrated synthetic biological systems has been published. They have built on previously well-characterized dynamic modules as well as parts. Top-down decomposition and bottom-up assembly have allowed such reuse.
-
As the field moves forward, part standardization and computational design tools are likely to make the design and implementation of regulatory networks more predictable. New classes of sensors for chemical or optic stimuli, and new classes of actuators such as master regulators of mammalian cellular processes will broaden the scope of synthetic biology.
-
Interactions with the cellular and extracellular context, nongenetic phenomena such as the sensing and actuation of mechanical forces, and complex nonlinear interactions will require strategies for orthogonalization and insulation and may benefit from combining rational design with library selections.
Abstract
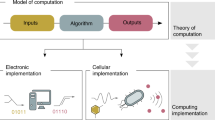
Synthetic gene circuits are designed to program new biological behaviour, dynamics and logic control. For all but the simplest synthetic phenotypes, this requires a structured approach to map the desired functionality to available molecular and cellular parts and processes. In other engineering disciplines, a formalized design process has greatly enhanced the scope and rate of success of projects. When engineering biological systems, a desired function must be achieved in a context that is incompletely known, is influenced by stochastic fluctuations and is capable of rich nonlinear interactions with the engineered circuitry. Here, we review progress in the provision and engineering of libraries of parts and devices, their composition into large systems and the emergence of a formal design process for synthetic biology.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Gardner, T. S., Cantor, C. R. & Collins, J. J. Construction of a genetic toggle switch in Escherichia coli. Nature 403, 339–342 (2000).
Weiss, R. & Basu, S. The device physics of cellular logic gates. in NSC-1: The First Workshop on Non-Silicon Computing 54–61 (2002).
Elowitz, M. B. & Leibler, S. A synthetic oscillatory network of transcriptional regulators. Nature 403, 335–338 (2000).
Mukherji, S. & van Oudenaarden, A. Synthetic biology: understanding biological design from synthetic circuits. Nature Rev. Genet. 10, 859–871 (2009).
Nandagopal, N. & Elowitz, M. B. Synthetic biology: integrated gene circuits. Science 333, 1244–1248 (2011).
Ruder, W. C., Lu, T. & Collins, J. J. Synthetic biology moving into the clinic. Science 333, 1248–1252 (2011).
Khalil, A. S. & Collins, J. J. Synthetic biology: applications come of age. Nature Rev. Genet. 11, 367–379 (2010).
Yizhar, O. et al. Neocortical excitation/inhibition balance in information processing and social dysfunction. Nature 477, 171–178 (2011).
Carr, P. A. & Church, G. M. Genome engineering. Nature Biotech. 27, 1151–1162 (2009).
Ellis, T., Adie, T. & Baldwin, G. S. DNA assembly for synthetic biology: from parts to pathways and beyond. Integr. Biol. 3, 109–118 (2011).
Ro, D.-K. et al. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature 440, 940–943 (2006).
Wang, H. H. et al. Programming cells by multiplex genome engineering and accelerated evolution. Nature 460, 894–898 (2009). Optimization of a gene network by simultaneous modification of multiple ribosome binding sites across a bacterial genome is discussed in this paper. It also shows the potential of fast and efficient genome engineering.
Weber, W. et al. A synthetic mammalian gene circuit reveals antituberculosis compounds. Proc. Natl Acad. Sci. USA 105, 9994–9998 (2008).
Purnick, P. E. M. & Weiss, R. The second wave of synthetic biology: from modules to systems. Nature Rev. Mol. Cell Biol. 10, 410–422 (2009).
Todd, M. H. Computer-aided organic synthesis. Chem. Soc. Rev. 34, 247–266 (2005).
MacDonald, J. T., Barnes, C., Kitney, R. I., Freemont, P. S. & Stan, G.-B. V. Computational design approaches and tools for synthetic biology. Integr. Biol. 3, 97–108 (2011).
Chandran, D., Bergmann, F. T., Sauro, H. M. & Densmore, D. Design and Analysis of Biomolecular Circuits: Engineering Approaches to Systems and Synthetic Biology 203–224 (Springer, 2011).
Beal, J., Lu, T. & Weiss, R. Automatic compilation from high-level biologically-oriented programming language to genetic regulatory networks. PLoS ONE 6, e22490 (2011).
Grünberg, R. & Serrano, L. Strategies for protein synthetic biology. Nucleic Acids Res. 38, 2663–2675 (2010).
Martin, A. R. C. et al. Protein folds and functions. Structure 6, 875–884 (1998).
Lutz, R. & Bujard, H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 25, 1203–1210 (1997).
Choo, Y., Sánchez-García, I. & Klug, A. In vivo repression by a site-specific DNA-binding protein designed against an oncogenic sequence. Nature 372, 642–645 (1994).
Klug, A. The discovery of zinc fingers and their applications in gene regulation and genome manipulation. Annu. Rev. Biochem. 79, 213–231 (2010).
Boch, J. et al. Breaking the code of DNA binding specificity of TAL-type III effectors. Science 326, 1509–1512 (2009).
Moscou, M. J. & Bogdanove, A. J. A simple cipher governs DNA recognition by TAL effectors. Science 326, 1501 (2009).
Voytas, D. F. & Joung, J. K. D. N. A. Binding made easy. Science 326, 1491–1492 (2009).
Morbitzer, R., Römer, P., Boch, J. & Lahaye, T. Regulation of selected genome loci using de novo-engineered transcription activator-like effector (TALE)-type transcription factors. Proc. Natl Acad. Sci. USA 107, 1–6 (2010).
Davidson, E. A. & Ellington, A. D. Synthetic RNA circuits. Nature Chem. Biol. 3, 23–28 (2007).
Isaacs, F. J., Dwyer, D. J. & Collins, J. J. RNA synthetic biology. Nature Biotech. 24, 545–554 (2006).
Salis, H. M. Mirsky, E. A. & Voigt, C. A. Automated design of synthetic ribosome binding sites to control protein expression. Nature Biotech. 27, 946–950 (2009). This study uses a physical chemical model of the interaction between the Shine–Dalgarno sequence and the 16S ribosomal RNA for predictive forward design of ribosomal binding sites of desired strength.
Yokobayashi, Y., Weiss, R. & Arnold, F. H. Directed evolution of a genetic circuit. Proc. Natl Acad. Sci. USA 99, 16587–16591 (2002).
Deans, T. L., Cantor, C. R. & Collins, J. J. A tunable genetic switch based on RNAi and repressor proteins for regulating gene expression in mammalian cells. Cell 130, 363–372 (2007).
Rinaudo, K. et al. A universal RNAi-based logic evaluator that operates in mammalian cells. Nature Biotech. 25, 795–801 (2007).
Xie, Z., Liu, S. J., Bleris, L. & Benenson, Y. Logic integration of mRNA signals by an RNAi-based molecular computer. Nucleic Acids Res. 38, 2692–2701 (2010).
Xie, Z., Wroblewska, L., Prochazka, L., Weiss, R. & Benenson, Y. Multi-input RNAi-based logic circuit for identification of specific cancer cells. Science 333, 1307–1311 (2011). This paper shows that the use of multiple miRNA biomarker sensors and synthetic genetic logic for the specific identification of a particular human cancer cell type.
Lucks, J. B., Qi, L., Mutalik, V. K., Wang, D. & Arkin, A. P. Versatile RNA-sensing transcriptional regulators for engineering genetic networks. Proc. Natl Acad. Sci. USA 108, 8617–8622 (2011).
Cho, E. J., Lee, J.-W. & Ellington, A. D. Applications of aptamers as sensors. Annu. Rev. Anal. Chem. 2, 241–264 (2009).
Famulok, M., Hartig, J. S. & Mayer, G. Functional aptamers and aptazymes in biotechnology, diagnostics, and therapy. Chem. Rev. 107, 3715–3743 (2007).
Win, M. N. & Smolke, C. D. A modular and extensible RNA-based gene-regulatory platform for engineering cellular function. Proc. Natl Acad. Sci. USA 104, 14283–14288 (2007).
Win, M. N. & Smolke, C. D. Higher-order cellular information processing with synthetic RNA devices. Science 322, 456–460 (2008).
Ausländer, S., Ketzer, P. & Hartig, J. S. A ligand-dependent hammerhead ribozyme switch for controlling mammalian gene expression. Mol. Biosyst. 6, 807–814 (2010).
Joyce, G. F. Forty years of in vitro evolution. Angew. Chem. Int. Edn Engl. 46, 6420–6436 (2007).
Culler, S. J., Hoff, K. G. & Smolke, C. D. Reprogramming cellular behavior with RNA controllers responsive to endogenous proteins. Science 330, 1251–1255 (2010).
Vuyisich, M. & Beal, P. A. Controlling protein activity with ligand-regulated RNA aptamers. Chem. Biol. 9, 907–913 (2002).
Hunsicker, A. et al. An RNA aptamer that induces transcription. Chem. Biol. 16, 173–180 (2009).
Skerker, J. M. et al. Rewiring the specificity of two-component signal transduction systems. Cell 133, 1043–1054 (2008).
Levskaya, A. et al. Synthetic biology: engineering Escherichia coli to see light. Nature 438, 441–442 (2005).
Tabor, J. J. Levskaya, A. & Voigt, C. A. Multichromatic control of gene expression in Escherichia coli. J. Mol. Biol. 405, 315–324 (2010).
Toettcher, J. E., Voigt, C. A., Weiner, O. D. & Lim, W. A. The promise of optogenetics in cell biology: interrogating molecular circuits in space and time. Nature Methods 8, 35–38 (2011).
Fenno, L., Yizhar, O. & Deisseroth, K. The development and application of optogenetics. Annu. Rev. Neurosci. 34, 389–412 (2010).
Boyden, E. S., Zhang, F., Bamberg, E., Nagel, G. & Deisseroth, K. Millisecond-timescale, genetically targeted optical control of neural activity. Nature Neurosci. 8, 1263–1268 (2005).
Levskaya, A. Weiner, O. D., Lim, W. A. & Voigt, C. A. Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature 461, 997–1001 (2009).
Wu, Y. I. et al. A genetically encoded photoactivatable Rac controls the motility of living cells. Nature 461, 104–108 (2009).
Airan, R. D., Thompson, K. R., Fenno, L. E., Bernstein, H. & Deisseroth, K. Temporally precise in vivo control of intracellular signalling. Nature 458, 1025–1029 (2009).
Gautier, A., Deiters, A. & Chin, J. W. Light-activated kinases enable temporal dissection of signaling networks in living cells. J. Am. Chem. Soc. 133, 2124–2127 (2011).
Magnus, C. J. et al. Chemical and genetic engineering of selective ion channel-ligand interactions. Science 333, 1292–1296 (2011).
Pei, Y., Rogan, S. C., Yan, F. & Roth, B. L. Engineered GPCRs as tools to modulate signal transduction. Physiology 23, 313–321 (2008).
Dong, S., Rogan, S. C. & Roth, B. L. Directed molecular evolution of DREADDs: a generic approach to creating next-generation RASSLs. Nature Protoc. 5, 561–573 (2010).
Lim, W. A. Designing customized cell signalling circuits. Nature Rev. Mol. Cell Biol. 11, 393–403 (2010). This paper reviews a series of studies conducted in the Lim group on engineering the dynamics of protein–protein interaction networks in eukaryotic signal processing by protein domain recombination. Although challenging, this is an important complement to the more widespread engineering of transcriptional regulation.
Burrill, D. R. & Silver, P. A. Making cellular memories. Cell 140, 13–18 (2010).
Purcell, O., Savery, N. J., Grierson, C. S. & di Bernardo, M. A comparative analysis of synthetic genetic oscillators. J. R. Soc. Interface 7, 1503–1524 (2010).
Atkinson, M. R., Savageau, M. A., Myers, J. T. & Ninfa, A. J. Development of genetic circuitry exhibiting toggle switch or oscillatory behavior in Escherichia coli. Cell 113, 597–607 (2003).
Stricker, J. et al. A fast, robust and tunable synthetic gene oscillator. Nature 456, 516–519 (2008).
Danino, T., Mondragón-Palomino, O., Tsimring, L. & Hasty, J. A synchronized quorum of genetic clocks. Nature 463, 326–330 (2010).
Tigges, M., Marquez-Lago, T. T., Stelling, J. & Fussenegger, M. A tunable synthetic mammalian oscillator. Nature 457, 309–312 (2009).
Kramer, B. P. et al. An engineered epigenetic transgene switch in mammalian cells. Nature Biotech. 22, 867–870 (2004).
Ham, T. S., Lee, S. K., Keasling, J. D. & Arkin, A. P. Design and construction of a double inversion recombination switch for heritable sequential genetic memory. PLoS ONE 3, e2815 (2008).
Friedland, A. E. et al. Synthetic gene networks that count. Science 324, 1199–1202 (2009).
Brenner, K., You, L. & Arnold, F. H. Engineering microbial consortia: a new frontier in synthetic biology. Trends Biotechnol. 26, 483–489 (2008).
Pai, A., Tanouchi, Y., Collins, C. H. & You, L. Engineering multicellular systems by cell-cell communication. Curr. Opin. Biotechnol. 20, 461–470 (2009).
Basu, S., Gerchman, Y., Collins, C. H., Arnold, F. H. & Weiss, R. A synthetic multicellular system for programmed pattern formation. Nature 434, 1130–1134 (2005).
Liu, C. et al. Sequential establishment of stripe patterns in an expanding cell population. Science 334, 238–241 (2011).
You, L., Cox, R. S., Weiss, R. & Arnold, F. H. Programmed population control by cell-cell communication and regulated killing. Nature 428, 868–871 (2004).
Balagaddé, F. K. et al. A synthetic Escherichia coli predator-prey ecosystem. Mol. Systems Biol. 4, 187 (2008).
Weber, W., Daoud-El Baba, M. & Fussenegger, M. Synthetic ecosystems based on airborne inter- and intrakingdom communication. Proc. Natl Acad. Sci. USA 104, 10435–10440 (2007).
Tamsir, A. Tabor, J. J. & Voigt, C. A. Robust multicellular computing using genetically encoded NOR gates and chemical “wires”. Nature 469, 212–215 (2011). References 76 and 100 demonstrate the decomposition of complex biological logic and dynamics into elementary functions which are implemented in single cells and composed via cell–cell communication in a population.
Tabor, J. J. et al. A synthetic genetic edge detection program. Cell 137, 1272–1281 (2009). An integrated system is described in this paper that combines light sensing, photographic inversion and cell–cell communication modules to produce a pigment only along the edges between illuminated and non-illuminated areas of a bacterial culture on solid medium.
Collins, C. H., Leadbetter, J. R. & Arnold, F. H. Dual selection enhances the signaling specificity of a variant of the quorum-sensing transcriptional activator LuxR. Nature Biotech. 24, 708–712 (2006).
Sturme, M. H. J. et al. Cell to cell communication by autoinducing peptides in gram-positive bacteria. Antonie van Leeuwenhoek 81, 233–243 (2002).
Dunny, G. M. & Leonard, B. A. Cell–cell communication in Gram-positive bacteria. Annu. Rev. Microbiol. 51, 527–564 (1997).
Clarke, E. J. & Voigt, C. A. Characterization of combinatorial patterns generated by multiple two-component sensors in E. coli that respond to many stimuli. Biotechnol. Bioeng. 108, 666–675 (2011).
Ninfa, A. J. Use of two-component signal transduction systems in the construction of synthetic genetic networks. Curr. Opin. Microbiol. 13, 240–245 (2010).
Shou, W., Ram, S. & Vilar, J. M. G. Synthetic cooperation in engineered yeast populations. Proc. Natl Acad. Sci. USA 104, 1877–1882 (2007).
Weber, W., Schuetz, M., Dénervaud, N. & Fussenegger, M. A synthetic metabolite-based mammalian inter-cell signaling system. Mol. Biosyst. 5, 757–763 (2009).
Weber, W. et al. Gas-inducible transgene expression in mammalian cells and mice. Nature Biotech. 22, 1440–1444 (2004).
Wang, W.-D., Chen, Z.-T., Kang, B.-G. & Li, R. Construction of an artificial intercellular communication network using the nitric oxide signaling elements in mammalian cells. Exp. Cell Res. 314, 699–706 (2008).
Chen, M.-T. & Weiss, R. Artificial cell–cell communication in yeast Saccharomyces cerevisiae using signaling elements from Arabidopsis thaliana. Nature Biotech. 23, 1551–1555 (2005).
Park, S.-H. Zarrinpar, A. & Lim, W. A. Rewiring MAP kinase pathways using alternative scaffold assembly mechanisms. Science 299, 1061–1064 (2003).
Dueber, J. E. Mirsky, E. A. & Lim, W. A. Engineering synthetic signaling proteins with ultrasensitive input/output control. Nature Biotech. 25, 660–662 (2007).
Bashor, C. J. Helman, N. C., Yan, S. & Lim, W. A. Using engineered scaffold interactions to reshape MAP kinase pathway signaling dynamics. Science 319, 1539–1543 (2008).
Peisajovich, S. G., Garbarino, J. E., Wei, P. & Lim, W. A. Rapid diversification of cell signaling phenotypes by modular domain recombination. Science 328, 368–372 (2010).
Alon, U. Network motifs: theory and experimental approaches. Nature Rev. Genet. 8, 450–461 (2007).
Guet, C. C., Elowitz, M. B., Hsing, W. & Leibler, S. Combinatorial synthesis of genetic networks. Science 296, 1466–1470 (2002).
Francois, P., Hakim, V. & Siggia, E. D. Deriving structure from evolution: metazoan segmentation. Mol. Syst. Biol. 3, 154 (2007).
François, P. & Hakim, V. Design of genetic networks with specified functions by evolution in silico. Proc. Natl Acad. Sci. USA 101, 580–585 (2004).
Ellis, T., Wang, X. & Collins, J. J. Diversity-based, model-guided construction of synthetic gene networks with predicted functions. Nature Biotech. 27, 465–471 (2009). This study achieved predictive, systems-level design of sophisticated regulatory dynamics by quantitative experimental characterization of a library of ribosomal binding sites and computational system design.
Randall, A., Guye, P., Gupta, S., Duportet, X. & Weiss, R. Design and connection of robust genetic circuits. Meth. Enzymol. 497, 159–186 (2011).
Silva-Rocha, R. & de Lorenzo, V. Noise and robustness in prokaryotic regulatory networks. Annu. Rev. Microbiol. 64, 257–275 (2010).
Balázsi, G., van Oudenaarden, A. & Collins, J. J. Cellular decision making and biological noise: from microbes to mammals. Cell 144, 910–925 (2011).
Regot, S. et al. Distributed biological computation with multicellular engineered networks. Nature 469, 207–211 (2011). See the blurb for reference 76.
Del Vecchio, D., Ninfa, A. J. & Sontag, E. D. Modular cell biology: retroactivity and insulation. Mol. Systems Biol. 4, 161 (2008). This paper derives a model of retroactivity, whereby downstream modules can alter upstream dynamics — for example, by sequestration effects — and proposes several potential insulation mechanisms to minimize retroactivity.
Mukherji, S. et al. MicroRNAs can generate thresholds in target gene expression. Nature Genet. 43, 854–859 (2011).
Nandagopal, N. & Elowitz, M. B. Synthetic biology: integrated gene circuits. Science 333, 1244–1248 (2011).
Ye, H., Daoud-El Baba, M., Peng, R.-W. & Fussenegger, M. A synthetic optogenetic transcription device enhances blood-glucose homeostasis in mice. Science 332, 1565–1568 (2011). This paper provides an integrated synthetic gene circuit using both synthetic and endogenous modules for a proof-of-concept of a potential gene or cell-based synthetic biomedical therapy.
Anderson, J. C. Clarke, E. J., Arkin, A. P. & Voigt, C. A. Environmentally controlled invasion of cancer cells by engineered bacteria. J. Mol. Biol. 355, 619–627 (2006).
Haseltine, E. L. & Arnold, F. H. Synthetic gene circuits: design with directed evolution. Annu. Rev. Biophys. Biomol. Struct. 36, 1–19 (2007).
Röthlisberger, D. et al. Kemp elimination catalysts by computational enzyme design. Nature 453, 190–195 (2008).
Voigt, C. A., Mayo, S. L., Arnold, F. H. & Wang, Z. G. Computational method to reduce the search space for directed protein evolution. Proc. Natl Acad. Sci. USA 98, 3778–3783 (2001).
Lutz, S. & Patrick, W. M. Novel methods for directed evolution of enzymes: quality, not quantity. Curr. Opin. Biotechnol. 15, 291–297 (2004).
Katz, R. H. Contemporary Logic Design. (Benjamin Cummings, 1994).
Corey, E. J. The logic of chemical synthesis: multistep synthesis of complex carbogenic molecules (Nobel Lecture). Angew. Chem. Int. Edn Engl. 30, 455–465 (1991).
Corey, E., Long, A. & Rubenstein, S. Computer-assisted analysis in organic synthesis. Science 228, 408–418 (1985).
Hoogenboom, H. R. Selecting and screening recombinant antibody libraries. Nature Biotech. 23, 1105–1116 (2005).
Hackel, B. J., Kapila, A. & Wittrup, K. D. Picomolar affinity fibronectin domains engineered utilizing loop length diversity, recursive mutagenesis, and loop shuffling. J. Mol. Biol. 381, 1238–1252 (2008).
Boersma, Y. L. & Plückthun, A. DARPins and other repeat protein scaffolds: advances in engineering and applications. Curr. Opin. Biotechnol. 22, 849–57 (2011).
Leisner, M., Bleris, L., Lohmueller, J., Xie, Z. & Benenson, Y. Rationally designed logic integration of regulatory signals in mammalian cells. Nature Nanotechnol. 5, 1–5 (2010).
Cermak, T. et al. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 39, e82 (2011).
Miller, J. C. et al. A TALE nuclease architecture for efficient genome editing. Nature Biotech. 29, 143–148 (2011).
Conklin, B. R. et al. Engineering GPCR signaling pathways with RASSLs. Persp. 5, 673–678 (2008).
Gunaydin, L. a. et al. Ultrafast optogenetic control. Nature Neurosci. 13, 387–392 (2010).
Weissman, K. J. & Leadlay, P. F. Combinatorial biosynthesis of reduced polyketides. Nature Rev. Microbiol. 3, 925–936 (2005).
Cane, D. E. Harnessing the biosynthetic code: combinations, permutations, and mutations. Science 282, 63–68 (1998).
Jiang, L. et al. De novo computational design of retro-aldol enzymes. Science 319, 1387–1391 (2008).
Richter, F., Leaver-Fay, A., Khare, S. D., Bjelic, S. & Baker, D. De novo enzyme design using Rosetta3. PLoS ONE 6, e19230 (2011).
Kramer, B. P., Fischer, C. & Fussenegger, M. BioLogic gates enable logical transcription control in mammalian cells. Biotechnol. Bioeng. 87, 478–484 (2004).
Tigges, M., Dénervaud, N., Greber, D., Stelling, J. & Fussenegger, M. A synthetic low-frequency mammalian oscillator. Nucleic Acids Res. 38, 2702–2711 (2010).
Swinburne, I., Miguez, D. G., Landgraf, D. & Silver, P. Intron length increases oscillatory periods of gene expression in animal cells. Genes Dev. 22, 2342–2346 (2008).
Gerhart, J. & Kirschner, M. The theory of facilitated variation. Proc. Natl Acad. Sci. USA 104, 8582–8589 (2007).
Yuh, C. H., Bolouri, H. & Davidson, E. H. Genomic cis-regulatory logic: experimental and computational analysis of a sea urchin gene. Science 279, 1896–1902 (1998).
Green, J. & Langer, R. A combinatorial polymer library approach yields insight into nonviral gene delivery. Acc. Chem. Res. 41, 749–759 (2008).
Kauffman, S. A. The Origins of Order: Self-Organization and Selection in Evolution (Oxford Univ. Press, 1993).
Kauffman, S. A. & Weinberger, E. D. The NK model of rugged fitness landscapes and its application to maturation of the immune response. J. Theor. Biol. 141, 211–245 (1989).
Funahashi, A. et al. CellDesigner 3.5: a versatile modeling tool for biochemical networks. Proc. IEEE 96, 1254–1265 (2008).
Pedersen, M. & Phillips, A. Towards programming languages for genetic engineering of living cells. J. R. Soc. Interface 6, S437–S450 (2009).
Czar, M. J., Cai, Y. & Peccoud, J. Writing DNA with GenoCAD. Nucleic Acids Res. 37, W40–W47 (2009).
Mirschel, S., Steinmetz, K., Rempel, M. & Ginkel, M. PROMOT: modular modeling for systems biology. Bioinformatics 25, 687–689 (2009).
Hill, A. D., Tomshine, J. R., Weeding, E. M. B., Sotiropoulos, V. & Kaznessis, Y. N. SynBioSS: the synthetic biology modeling suite. Bioinformatics 24, 2551–2553 (2008).
Chandran, D., Bergmann, F. T. & Sauro, H. M. TinkerCell: modular CAD tool for synthetic biology. J. Biol. Eng. 29, 19 (2009).
Rodrigo, G., Carrera, J. & Jaramillo, A. Genetdes: automatic design of transcriptional networks. Bioinformatics 23, 1857–1858 (2007). (2009).
Dasika, M. S. & Maranas, C. D. OptCircuit: an optimization based method for computational design of genetic circuits. BMC Systems Biol. 2, 24 (2008).
Batt, G., Yordanov, B., Weiss, R. & Belta, C. Robustness analysis and tuning of synthetic gene networks. Bioinformatics 23, 2415–2422 (2007).
Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31, 3406–3415 (2003).
Hoops, S. et al. COPASI-a COmplex PAthway SImulator. Bioinformatics 22, 3067–3074 (2006).
Merks, R. & Glazier, J. A cell-centered approach to developmental biology. Physica A 352, 113–130 (2005).
Villalobos, A., Ness, J. E., Gustafsson, C., Minshull, J. & Govindarajan, S. Gene Designer: a synthetic biology tool for constructing artificial DNA segments. BMC Bioinformat. 7, 285 (2006).
Richardson, S. M., Wheelan, S. J., Yarrington, R. M. & Boeke, J. D. GeneDesign: rapid, automated design of multikilobase synthetic genes. Genome Res. 16, 550–556 (2006).
Xia, B. et al. Developer's and user's guide to Clotho v2.0 A software platform for the creation of synthetic biological systems. Meth. Enzymol. 498, 97–135 (2011).
Acknowledgements
A.L.S. is pleased to thank the Boehringer Ingelheim Fonds for support through a Ph.D. fellowship. Work in the Weiss laboratory is supported by the US Defense Advanced Research Projects Agency, the US National Institutes of Health and the US National Science Foundation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Glossary
- Abstraction
-
The process of hiding the extraneous details of a specific implementation to highlight the salient and general features of a system or design.
- Actuation
-
The action on the internal or external environment that constitutes the output of a synthetic gene circuit.
- TIM barrel
-
A conserved protein fold named after triose phosphate isomerase (TIM) and shared among many enzymes with widely differing substrate specificities and catalytic activities.
- Immunoglobulin fold
-
A very common protein fold that is based on a β-sandwich. Contains hypervariable loops, which can accommodate almost any sequence and bind a wide variety of partners.
- Photocaged unnatural amino acids
-
Unnatural amino acids containing a photosensitive masking group, which following activation by light reveals a biologically active functional group.
- Quorum sensing
-
Sensing of population density by cell–cell communication.
- Oscillators
-
A circuit with a periodically varying output signal.
- Bandpass filters
-
A circuit that lets through signals within a certain frequency range but not outside it.
- Topology
-
In a network, the set of all connections among nodes. Depending on what the network signifies (for example, molecular binding, genetic regulation or metabolic fluxes), the network topology takes different meanings. For synthetic gene circuits, topology usually refers to regulatory relationships.
- Two-component signalling systems
-
A type of response system commonly found in bacteria and typically consisting of a membrane-bound, sensory histidine kinase and a soluble response regulator.
- Signal transduction
-
The triggering of an intracellular event following detection of an extracellular cue by a transmembrane receptor molecule.
- NAND gate
-
A digital logic gate that implements the logical NAND, or 'NOT AND'. Its output is low when all inputs are high and is otherwise high.
- NOR gates
-
A digital logic gate that implements the logical NOR, or 'NOT OR'. Its output is low when at least one input is high and is otherwise high.
- AND gates
-
Digital logic gates that implement the logical AND. Their output is high when all inputs are high and is otherwise low.
- Binary addition with carry
-
Addition of numbers represented in a base-2 numeral system, where care is taken to carry digits to the left as necessary. For example, 01b + 01b = 10b (in decimal numbers, 1 + 1 = 2).
- Emergent
-
A term used to describe a phenomenon whereby a system is more than the sum of its parts. An emergent property or behaviour is irreducible.
- Kinetic parameters
-
In a mass action kinetic model of biological dynamics, the kinetic parameters are the constants in the differential equations governing the dynamics of a system, such as rate constants and Hill coefficients.
Rights and permissions
About this article
Cite this article
Slusarczyk, A., Lin, A. & Weiss, R. Foundations for the design and implementation of synthetic genetic circuits. Nat Rev Genet 13, 406–420 (2012). https://doi.org/10.1038/nrg3227
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrg3227
This article is cited by
-
Synthesizing cellular LOGIC
Nature Chemical Biology (2023)
-
Applications of synthetic biology in medical and pharmaceutical fields
Signal Transduction and Targeted Therapy (2023)
-
2D printed multicellular devices performing digital and analogue computation
Nature Communications (2021)
-
Engineering living therapeutics with synthetic biology
Nature Reviews Drug Discovery (2021)
-
A biological multiplexer, designs, and simulations
The Journal of Supercomputing (2021)