Abstract
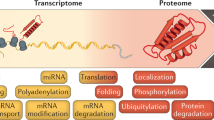
Recent advances in next-generation DNA sequencing and proteomics provide an unprecedented ability to survey mRNA and protein abundances. Such proteome-wide surveys are illuminating the extent to which different aspects of gene expression help to regulate cellular protein abundances. Current data demonstrate a substantial role for regulatory processes occurring after mRNA is made — that is, post-transcriptional, translational and protein degradation regulation — in controlling steady-state protein abundances. Intriguing observations are also emerging in relation to cells following perturbation, single-cell studies and the apparent evolutionary conservation of protein and mRNA abundances. Here, we summarize current understanding of the major factors regulating protein expression.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
de Sousa Abreu, R., Penalva, L. O., Marcotte, E. & Vogel, C. Global signatures of protein and mRNA expression levels. Mol. Biosyst. 5, 1512–1526 (2009).
Hu, Q. et al. The Orbitrap: a new mass spectrometer. J. Mass Spectrom. 40, 430–443 (2005).
Mallick, P. & Kuster, B. Proteomics: a pragmatic perspective. Nature Biotech. 28, 695–709 (2010).
Lu, P., Vogel, C., Wang, R., Yao, X. & Marcotte, E. M. Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nature Biotech. 25, 117–124 (2007).
Schwanhäusser, B. et al. Global quantification of mammalian gene expression control. Nature 473, 337–342 (2011).
Malmstrom, J. et al. Proteome-wide cellular protein concentrations of the human pathogen Leptospira interrogans. Nature 460, 762–765 (2009).
Gerber, S. A., Rush, J., Stemman, O., Kirschner, M. W. & Gygi, S. P. Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc. Natl Acad. Sci. USA 100, 6940–6945 (2003).
Ong, S. E. et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 (2002).
Ross, P. L. et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteomics 3, 1154–1169 (2004).
Bantscheff, M., Schirle, M., Sweetman, G., Rick, J. & Kuster, B. Quantitative mass spectrometry in proteomics: a critical review. Anal. Bioanal. Chem. 389, 1017–1031 (2007).
Martin, J. A. & Wang, Z. Next-generation transcriptome assembly. Nature Rev. Genet. 12, 671–682 (2011).
Ingolia, N. T., Ghaemmaghami, S., Newman, J. R. & Weissman, J. S. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218–223 (2009).
Taniguchi, Y. et al. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science 329, 533–538 (2010).
Newman, J. R. et al. Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature 441, 840–846 (2006).
Ghaemmaghami, S. et al. Global analysis of protein expression in yeast. Nature 425, 737–741 (2003).
Belle, A., Tanay, A., Bitincka, L., Shamir, R. & O'Shea, E. K. Quantification of protein half-lives in the budding yeast proteome. Proc. Natl Acad. Sci. USA 103, 13004–13009 (2006).
Eden, E. et al. Proteome half-life dynamics in living human cells. Science 331, 764–768 (2011).
Yen, H. C., Xu, Q., Chou, D. M., Zhao, Z. & Elledge, S. J. Global protein stability profiling in mammalian cells. Science 322, 918–923 (2008).
Maier, T., Guell, M. & Serrano, L. Correlation of mRNA and protein in complex biological samples. FEBS Lett. 583, 3966–3973 (2009).
de Godoy, L. M. et al. Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature 455, 1251–1254 (2008).
Lundberg, E. et al. Defining the transcriptome and proteome in three functionally different human cell lines. Mol. Syst. Biol. 6, 450 (2010).
Sharova, L. V. et al. Database for mRNA half-life of 19 977 genes obtained by DNA microarray analysis of pluripotent and differentiating mouse embryonic stem cells. DNA Res. 16, 45–58 (2009).
Selinger, D. W., Saxena, R. M., Cheung, K. J., Church, G. M. & Rosenow, C. Global RNA half-life analysis in Escherichia coli reveals positional patterns of transcript degradation. Genome Res. 13, 216–223 (2003).
Vogel, C. et al. Sequence signatures and mRNA concentration can explain two-thirds of protein abundance variation in a human cell line. Mol. Syst. Biol. 6, 400 (2010).
Lee, M. V. et al. A dynamic model of proteome changes reveals new roles for transcript alteration in yeast. Mol. Syst. Biol. 7, 514 (2011).
Fournier, M. L. et al. Delayed correlation of mRNA and protein expression in rapamycin-treated cells and a role for Ggc1 in cellular sensitivity to rapamycin. Mol. Cell. Proteomics 9, 271–284 (2009).
Vogel, C., Silva, G. M. & Marcotte, E. M. Protein expression regulation under oxidative stress. Mol. Cell. Proteomics 20 Sep 2011 (doi:10.1074/mcp.M111.009217).
Maier, T. et al. Quantification of mRNA and protein and integration with protein turnover in a bacterium. Mol. Syst. Biol. 7, 511 (2011).
Jayapal, K. P. et al. Uncovering genes with divergent mRNA-protein dynamics in Streptomyces coelicolor. PLoS ONE 3, e2097 (2008).
Li, G. W. & Xie, X. S. Central dogma at the single-molecule level in living cells. Nature 475, 308–315 (2011).
Fraser, H. B., Hirsh, A. E., Giaever, G., Kumm, J. & Eisen, M. B. Noise minimization in eukaryotic gene expression. PLoS Biol. 2, e137 (2004).
Gandhi, S. J., Zenklusen, D., Lionnet, T. & Singer, R. H. Transcription of functionally related constitutive genes is not coordinated. Nature Struct. Mol. Biol. 18, 27–34 (2011).
Pilpel, Y. Noise in biological systems: pros, cons, and mechanisms of control. Methods Mol. Biol. 759, 407–425 (2011).
Plotkin, J. B. Transcriptional regulation is only half the story. Mol. Syst. Biol. 6, 406 (2010).
Coghlan, A. & Wolfe, K. H. Relationship of codon bias to mRNA concentration and protein length in Saccharomyces cerevisiae. Yeast 16, 1131–1145 (2000).
Ingolia, N. T., Lareau, L. F. & Weissman, J. S. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of Mammalian proteomes. Cell 147, 789–802 (2011).
Mayr, C. & Bartel, D. P. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 138, 673–684 (2009).
Sandberg, R., Neilson, J. R., Sarma, A., Sharp, P. A. & Burge, C. B. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science 320, 1643–1647 (2008).
Schrimpf, S. P. et al. Comparative functional analysis of the Caenorhabditis elegans and Drosophila melanogaster proteomes. PLoS Biol. 7, e48 (2009).
Laurent, J. et al. Protein abundances are more conserved than mRNA abundances across diverse taxa. Proteomics 10, 4209–4212 (2010).
Weiss, M., Schrimpf, S., Hengartner, M. O., Lercher, M. J. & von Mering, C. Shotgun proteomics data from multiple organisms reveals remarkable quantitative conservation of the eukaryotic core proteome. Proteomics 10, 1297–1306 (2010).
Ramakrishnan, S. R. et al. Integrating shotgun proteomics and mRNA expression data to improve protein identification. Bioinformatics 25, 1397–1403 (2009).
Golding, I., Paulsson, J., Zawilski, S. M. & Cox, E. C. Real-time kinetics of gene activity in individual bacteria. Cell 123, 1025–1036 (2005).
Raj, A., Peskin, C. S., Tranchina, D., Vargas, D. Y. & Tyagi, S. Stochastic mRNA synthesis in mammalian cells. PLoS Biol. 4, e309 (2006).
Mukherji, S. et al. MicroRNAs can generate thresholds in target gene expression. Nature Genet. 43, 854–859 (2011).
Baek, D. et al. The impact of microRNAs on protein output. Nature 455, 64–71 (2008).
Selbach, M. et al. Widespread changes in protein synthesis induced by microRNAs. Nature 455, 58–63 (2008).
Jacobs, J. M. et al. Utilizing human blood plasma for proteomic biomarker discovery. J. Proteome Res. 4, 1073–1085 (2005).
Hebenstreit, D. et al. RNA sequencing reveals two major classes of gene expression levels in metazoan cells. Mol. Syst. Biol. 7, 497 (2011).
Dahan, O., Gingold, H. & Pilpel, Y. Regulatory mechanisms and networks couple the different phases of gene expression. Trends Genet. 27, 316–322 (2011).
Auld, K. L. & Silver, P. A. Transcriptional regulation by the proteasome as a mechanism for cellular protein homeostasis. Cell Cycle 5, 1503–1505 (2006).
Kodadek, T. No splicing, no dicing: non-proteolytic roles of the ubiquitin-proteasome system in transcription. J. Biol. Chem. 285, 2221–2226 (2010).
Acknowledgements
The authors acknowledge support from the US National Institutes of Health and the Welch Foundation (F1515, to E.M.M.). We also thank T. Lionnet for helpful discussions.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Glossary
- High-resolution tandem mass spectrometry
-
The use of two consecutive mass spectrometry steps to measure mass-to-charge ratios for peptides and their fragment ions, respectively. Modern technology enables a mass accuracy of <0.01 Da.
- Nanoflow chromatography
-
In the context of peptides, this method separates a peptide mixture by differences in biophysical properties. It operates at flow rates of nanolitres per minute to increase separation efficiency and decrease sample volumes.
- PEST sequences
-
Protein sequence motif enriched for proline (P), glutamate (E), serine (S) and threonine (T) that serves as a protein degradation signal.
- Ribosome footprinting
-
Identification of ribosomal binding sites on mRNA through ribosome stalling and next-generation sequencing of the bound RNA fragments.
- Stable isotopic labelling with amino acids in cell culture
-
(SILAC). A widely used technique for estimating relative protein concentrations by mass spectrometry.
Rights and permissions
About this article
Cite this article
Vogel, C., Marcotte, E. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet 13, 227–232 (2012). https://doi.org/10.1038/nrg3185
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrg3185
This article is cited by
-
Ribosome profiling: a powerful tool in oncological research
Biomarker Research (2024)
-
The glucocorticoid receptor as a master regulator of the Müller cell response to diabetic conditions in mice
Journal of Neuroinflammation (2024)
-
Benzo[a]pyrene disrupts LH/hCG-dependent mouse Leydig cell steroidogenesis through receptor/Gαs protein targeting
Scientific Reports (2024)
-
Drosophila eIF3f1 mediates host immune defense by targeting dTak1
EMBO Reports (2024)
-
Serpine1 mRNA confers mesenchymal characteristics to the cell and promotes CD8+ T cells exclusion from colon adenocarcinomas
Cell Death Discovery (2024)