Key Points
-
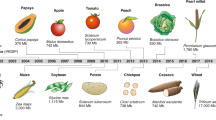
Reference genome sequences from many important crops and several model plant species are enabling a number of new applications of crop comparative genomics.
-
Compared with previous methods, comparative resequencing and high-density SNP genotyping permit much more detailed examination of crop evolutionary history and improve the potential to identify loci that are involved in plant domestication and improvement.
-
Genome-wide association studies and a new generation of genetic-mapping populations will improve assessment of the genetic basis of trait variation.
-
Deleterious mutations in individual genomes can be identified and selected against or even repaired.
-
Genomic selection or genome-wide marker-assisted selection can incorporate prior information on the effects of markers and accelerate plant breeding cycles.
Abstract
The completion of reference genome sequences for many important crops and the ability to perform high-throughput resequencing are providing opportunities for improving our understanding of the history of plant domestication and to accelerate crop improvement. Crop plant comparative genomics is being transformed by these data and a new generation of experimental and computational approaches. The future of crop improvement will be centred on comparisons of individual plant genomes, and some of the best opportunities may lie in using combinations of new genetic mapping strategies and evolutionary analyses to direct and optimize the discovery and use of genetic variation. Here we review such strategies and insights that are emerging.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Paterson, A. H., Freeling, M. & Sasaki, T. Grains of knowledge: genomics of model cereals. Genome Res. 15, 1643–1650 (2005).
Brown, A. H. D. Enzyme polymorphism in plant populations. Theor. Popul. Biol. 15, 1–42 (1979).
Bowers, J. E. et al. A high-density genetic recombination map of sequence-tagged sites for Sorghum, as a framework for comparative structural and evolutionary genomics of tropical grains and grasses. Genetics 165, 367–386 (2003).
Lewontin, R. C. & Krakauer, J. Distribution of gene frequency as a test of the theory of the selective neutrality of polymorphisms. Genetics 74, 175–195 (1973).
Nei, M. & Maruyama, T. Lewontin–Krakauer test for neutral genes — comment. Genetics 80, 395–395 (1975).
Schulte, D. et al. The International Barley Sequencing Consortium—at the threshold of efficient access to the barley genome. Plant Physiol. 149, 142–147 (2009).
Kaul, S. et al. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815 (2000).
Initiative, T. I. B. Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463, 763–768 (2010).
Pool, J. E., Hellmann, I., Jensen, J. D. & Nielsen, R. Population genetic inference from genomic sequence variation. Genome Res. 20, 291–300 (2010).
Metzker, M. L. Sequencing technologies — the next generation. Nature Rev. Genet. 11, 31–46 (2010).
Lockton, S. & Gaut, B. S. Plant conserved non-coding sequences and paralogue evolution. Trends Genet. 21, 60–65 (2005).
Haun, W. J. et al. The composition and origins of genomic variation among individuals of the soybean reference cultivar Williams 82. Plant Physiol. 155, 645–655 (2011).
Velasco, R. et al. A high quality draft consensus sequence of the genome of a heterozygous grapevine variety. PLoS ONE 2, e1326 (2007).
Gore, M. A. et al. A first-generation haplotype map of maize. Science 326, 1115–1117 (2009).
Young, N. D. et al. The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 16 Nov 2011(doi:1001038/nature10625).
Shulaev, V. et al. The genome of woodland strawberry (Fragaria vesca). Nature Genet. 43, 109–116 (2011).
Xu, X. et al. Genome sequence and analysis of the tuber crop potato. Nature 475, 189–195 (2011). This is an excellent example of both the challenges and promise of comparative genomics in crop plant genomes. To overcome polyploidy and high levels of heterozygosity, the authors use a combination of traditional Sanger and next-generation methods to sequence and annotate the genome of a doubled-monoploid potato line.
Rafalski, A. & Morgante, M. Corn and humans: recombination and linkage disequilibrium in two genomes of similar size. Trends Genet. 20, 103–111 (2004).
Lijavetzky, D., Cabezas, J. A., Ibanez, A., Rodriguez, V. & Martinez-Zapater, J. M. High throughput SNP discovery and genotyping in grapevine (Vitis vinifera L.) by combining a re-sequencing approach and SNPlex technology. BMC Genomics 8, 424 (2007).
Caldwell, K. S., Russell, J., Langridge, P. & Powell, W. Extreme population-dependent linkage disequilibrium detected in an inbreeding plant species, Hordeum vulgare. Genetics 172, 557–567 (2006).
Gaut, B. S. & Ross-Ibarra, J. Selection on major components of angiosperm genomes. Science 320, 484–486 (2008).
Tenaillon, M. I., Hollister, J. D. & Gaut, B. S. A triptych of the evolution of plant transposable elements. Trends Plant Sci. 15, 471–478 (2010).
Nordborg, M. & Weigel, D. Next-generation genetics in plants. Nature 456, 720–723 (2008).
Jaillon, O. et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449, 463–467 (2007).
Ross-Ibarra, J., Morrell, P. L. & Gaut, B. S. Colloquium papers: plant domestication, a unique opportunity to identify the genetic basis of adaptation. Proc. Natl Acad. Sci. USA 104 (Suppl. 1), 8641–8648 (2007).
Brown, A. H. D. Variation under domestication in plants: 1859 and today. Phil. Trans. R. Soc. B 365, 2523–2530 (2010).
Harris, D. R. Vavilov's concept of centres of origin of cultivated plants: its genesis and its influence on the study of agricultural origins. Biol. J. Linn. Soc. 39, 7–16 (1990).
Rosenberg, N. A. & Nordborg, M. Genealogical trees, coalescent theory and the analysis of genetic polymorphisms. Nature Rev. Genet. 3, 380–390 (2002). This is a very accessible introduction to genealogical histories and coalescent theory that are pertinent to interpretation of sequence polymorphism data.
Gaut, B. S. & Clegg, M. T. Molecular evolution of the Adh1 locus in the genus Zea. Proc. Natl Acad. Sci. USA 90, 5095–5099 (1993).
Kim, M. Y. et al. Whole-genome sequencing and intensive analysis of the undomesticated soybean (Glycine soja Sieb. and Zucc.) genome. Proc. Natl Acad. Sci. USA 107, 22032–22037 (2010).
Eyre-Walker, A., Gaut, R. L., Hilton, H., Feldman, D. L. & Gaut, B. S. Investigation of the bottleneck leading to the domestication of maize. Proc. Natl Acad. Sci. USA 95, 4441–4446 (1998).
Haudry, A. et al. Grinding up wheat: a massive loss of nucleotide diversity since domestication. Mol. Biol. Evol. 24, 1506–1517 (2007).
Wright, S. I. et al. The effects of artificial selection on the maize genome. Science 308, 1310–1314 (2005). This paper discusses a comparative resequencing study that used an original likelihood ratio test to model demography and selection. The paper was unique in providing an estimate of the proportion of loci in the genome involved in domestication and/or improvement.
Caicedo, A. L. et al. Genome-wide patterns of nucleotide polymorphism in domesticated rice. PLoS Genet. 3, 1745–1756 (2007).
Molina, J. et al. Molecular evidence for a single evolutionary origin of domesticated rice. Proc. Natl Acad. Sci. USA 108, 8351–8356 (2011).
Matsuoka, Y. et al. A single domestication for maize shown by multilocus microsatellite genotyping. Proc. Natl Acad. Sci. USA 99, 6080–6084 (2002).
Li, Y. H. et al. Genetic diversity in domesticated soybean (Glycine max) and its wild progenitor (Glycine soja) for simple sequence repeat and single-nucleotide polymorphism loci. New Phytol. 188, 242–253 (2010).
Chen, H., Morrell, P. L., Ashworth, V. E., de la Cruz, M. & Clegg, M. T. Tracing the geographic origins of major avocado cultivars. J. Hered. 100, 56–65 (2009).
Gepts, P. & Bliss, F. A. Phaseolin variability among wild and cultivated common beans (Phaseolus vulgaris) from Colombia. Econ. Bot. 40, 469–478 (1986).
Morrell, P. L. & Clegg, M. T. Genetic evidence for a second domestication of barley (Hordeum vulgare) east of the Fertile Crescent. Proc. Natl Acad. Sci. USA 104, 3289–3294 (2007).
Allaby, R. G., Fuller, D. Q. & Brown, T. A. The genetic expectations of a protracted model for the origins of domesticated crops. Proc. Natl Acad. Sci. USA 105, 13982–13986 (2008).
Ross-Ibarra, J. & Gaut, B. S. Multiple domestications do not appear monophyletic. Proc. Natl Acad. Sci. USA 105, E105; author reply E106 (2008).
Kwak, M. & Gepts, P. Structure of genetic diversity in the two major gene pools of common bean (Phaseolus vulgaris L., Fabaceae). Theor. Appl. Genet. 118, 979–992 (2009).
Myles, S. et al. Genetic structure and domestication history of the grape. Proc. Natl Acad. Sci. USA 108, 3530–3535 (2011).
van Heerwaarden, J. et al. Genetic signals of origin, spread, and introgression in a large sample of maize landraces. Proc. Natl Acad. Sci. USA 108, 1088–1092 (2011).
He, Z. et al. Two evolutionary histories in the genome of rice: the roles of domestication genes. PLoS Genet. 7, e1002100 (2011).
Price, A. L. et al. Sensitive detection of chromosomal segments of distinct ancestry in admixed populations. PLoS Genet. 5, e1000519 (2009).
Li, H. & Durbin, R. Inference of human population history from individual whole-genome sequences. Nature 475, 493–496 (2011).
Darwin, C. The Variation of Animals and Plants under Domestication (Appleton, New York, 1876).
Hammer, K. Das Domestikationssyndrom. Kulturpflanze 32, 11–34 (1984).
Burger, J. C., Chapman, M. A. & Burke, J. M. Molecular insights into the evolution of crop plants. Am. J. Bot. 95, 113–122 (2008).
Vigouroux, Y. et al. Identifying genes of agronomic importance in maize by screening microsatellites for evidence of selection during domestication. Proc. Natl Acad. Sci. USA 99, 9650–9655 (2002).
Chapman, M. A. et al. A genomic scan for selection reveals candidates for genes involved in the evolution of cultivated sunflower (Helianthus annuus). Plant Cell 20, 2931–2945 (2008).
Lam, H. M. et al. Resequencing of 31 wild and cultivated soybean genomes identifies patterns of genetic diversity and selection. Nature Genet. 42, 1053–1059 (2010).
Hurwitz, B. L. et al. Rice structural variation: a comparative analysis of structural variation between rice and three of its closest relatives in the genus Oryza. Plant J. 63, 990–1003 (2010).
Swanson-Wagner, R. A. et al. Pervasive gene content variation and copy number variation in maize and its undomesticated progenitor. Genome Res. 20, 1689–1699 (2010).
Buckler, E. S. et al. The genetic architecture of maize flowering time. Science 325, 714–718 (2009). This study, using the NAM population, found that flowering time in maize provides a good fit to classic models of a quantitative trait and that a large number of loci contribute additively to the phenotype.
Vielle-Calzada, J. P. et al. The Palomero genome suggests metal effects on domestication. Science 326, 1078–1078 (2009).
Sugimoto, K. et al. Molecular cloning of Sdr4, a regulator involved in seed dormancy and domestication of rice. Proc. Natl Acad. Sci. 107, 5792–5797 (2010).
Takahashi, R. The origin and evolution of cultivated barley. Adv. Genet. 7, 227–266 (1955).
Morrell, P. L. & Clegg, M. T. in Wild Crop Relatives: Genomic and Breeding Resources: Cereals (ed. Kole, C.) 309–320 (Springer, Berlin, 2011).
Jones, H. et al. Population-based resequencing reveals that the flowering time adaptation of cultivated barley originated east of the Fertile Crescent. Mol. Biol. Evol. 25, 2211–2219 (2008).
Bellon, M. R., Hodson, D. & Hellin, J. Assessing the vulnerability of traditional maize seed systems in Mexico to climate change. Proc. Natl Acad. Sci. USA 108, 13432–13437 (2011).
Komatsuda, T. et al. Six-rowed barley originated from a mutation in a homeodomain-leucine zipper I-class homeobox gene. Proc. Natl Acad. Sci. USA 104, 1424–1429 (2007).
Purugganan, M. D., Boyles, A. L. & Suddith, J. I. Variation and selection at the CAULIFLOWER floral homeotic gene accompanying the evolution of domesticated Brassica oleracea. Genetics 155, 855–862 (2000).
Teshima, K. M., Coop, G. & Przeworski, M. How reliable are empirical genomic scans for selective sweeps? Genome Res. 16, 702–712 (2006).
Ralph, P. & Coop, G. Parallel adaptation: one or many waves of advance of an advantageous allele? Genetics 186, 647–668 (2010).
Paterson, A. H. et al. Convergent domestication of cereal crops by independent mutations at corresponding genetic loci. Science 269, 1714–1718 (1995).
Goff, S. A. A unifying theory for general multigenic heterosis: energy efficiency, protein metabolism, and implications for molecular breeding. New Phytol. 189, 923–937 (2011).
Mauricio, R. Mapping quantitative trait loci in plants: uses and caveats for evolutionary biology. Nature Rev. Genet. 2, 370–381 (2001).
Risch, N. & Merikangas, K. The future of genetic studies of complex human diseases. Science 273, 1516–1517 (1996).
Myles, S. et al. Association mapping: critical considerations shift from genotyping to experimental design. Plant Cell 21, 2194–2202 (2009).
Mackay, T. F., Stone, E. A. & Ayroles, J. F. The genetics of quantitative traits: challenges and prospects. Nature Rev. Genet. 10, 565–577 (2009).
Wurschum, T. et al. Genome-wide association mapping of agronomic traits in sugar beet. Theor. Appl. Genet. 123, 1121–1131 (2011).
Saidou, A. A. et al. Association studies identify natural variation at PHYC linked to flowering time and morphological variation in pearl millet. Genetics 182, 899–910 (2009).
Huang, X. et al. Genome-wide association studies of 14 agronomic traits in rice landraces. Nature Genet. 42, 961–967 (2010).
Atwell, S. et al. Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature 465, 627–631 (2010).
Hamblin, M. T., Buckler, E. S. & Jannink, J. L. Population genetics of genomics-based crop improvement methods. Trends Genet. 27, 98–106 (2011).
Rockman, M. V. & Kruglyak, L. Breeding designs for recombinant inbred advanced intercross lines. Genetics 179, 1069–1078 (2008). This study provides an examination of breeding designs that maximize genetic resolution in intercross populations.
Macdonald, S. J. & Long, A. D. Joint estimates of quantitative trait locus effect and frequency using synthetic recombinant populations of Drosophila melanogaster. Genetics 176, 1261–1281 (2007). The authors of this paper provide a strong rationale for the development of next-generation populations. The study design permits estimation of QTL location, effect and frequency. Comparison of effect size of alleles contributed by founders of the population is particularly compelling.
Yu, J. M., Holland, J. B., McMullen, M. D. & Buckler, E. S. Genetic design and statistical power of nested association mapping in maize. Genetics 178, 539–551 (2008).
Brown, P. J. et al. Distinct genetic architectures for male and female inflorescence traits of maize. PLoS Genet. 7, e1002383 (2011).
Kump, K. L. et al. Genome-wide association study of quantitative resistance to southern leaf blight in the maize nested association mapping population. Nature Genet. 43, 163–168 (2011).
Tian, F. et al. Genome-wide association study of leaf architecture in the maize nested association mapping population. Nature Genet. 43, 159–162 (2011).
Cavanagh, C., Morell, M., Mackay, I. & Powell, W. From mutations to MAGIC: resources for gene discovery, validation and delivery in crop plants. Curr. Opin. Plant Biol. 11, 215–221 (2008).
Kover, P. X. et al. A multiparent advanced generation inter-cross to fine-map quantitative traits in Arabidopsis thaliana. PLoS Genet. 5, e1000551 (2009).
Harlan, H. V. & Martini, M. L. A composite hybrid mixture. J. Am. Soc. Agron. 487–490 (1929).
Suneson, C. A. An evolutionary plant breeding method. Agron. J. 48, 188–191 (1956).
Allard, R. W., Kahler, A. L. & Weir, B. S. The effect of selection on esterase allozymes in a barley population. Genetics 72, 489–503 (1972).
Clegg, M. T., Allard, R. W. & Kahler, A. L. Is the gene the unit of selection? Evidence from two experimental plant populations. Proc. Natl Acad. Sci. USA 69, 2474–2478 (1972).
Cargill, M. et al. Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nature Genet. 22, 231–238 (1999).
Manolio, T. A. et al. Finding the missing heritability of complex diseases. Nature 461, 747–753 (2009).
Johnson, T. & Barton, N. Theoretical models of selection and mutation on quantitative traits. Phil. Trans. R. Soc. Lond. B 360, 1411–1425 (2005).
Clark, A. G., Hubisz, M. J., Bustamante, C. D., Williamson, S. H. & Nielsen, R. Ascertainment bias in studies of human genome-wide polymorphism. Genome Res. 15, 1496–1502 (2005).
Altshuler, D. L. et al. A map of human genome variation from population-scale sequencing. Nature 467, 1061–1073 (2010).
Weigel, D. & Mott, R. The 1001 Genomes Project for Arabidopsis thaliana. Genome Biol. 10, 107 (2009).
Dickson, S. P., Wang, K., Krantz, I., Hakonarson, H. & Goldstein, D. B. Rare variants create synthetic genome-wide associations. PLoS Biol. 8, e1000294 (2010).
Hill, W. G. & Robertson, A. The effect of linkage on limits to artificial selection. Genet. Res. 8, 269–294 (1966).
Huff, C. D. et al. Crohn's disease and genetic hitchhiking at IBD5. Mol. Biol. Evol. 4 Aug 2011 (doi:10.1093/molbev/msr151).
Clegg, M. T. Measuring plant mating systems. Bioscience 30, 814–818 (1980).
Nielsen, R. Molecular signatures of natural selection. Annu. Rev. Genet. 39, 197–218 (2005).
Nielsen, R., Hellmann, I., Hubisz, M., Bustamante, C. & Clark, A. G. Recent and ongoing selection in the human genome. Nature Rev. Genet. 8, 857–868 (2007).
Walsh, B. Using molecular markers for detecting domestication, improvement, and adaptation genes. Euphytica 161, 1–17 (2008).
Asano, K. et al. Artificial selection for a green revolution gene during japonica rice domestication. Proc. Natl Acad. Sci. USA 108, 11034–11039 (2011).
Spielmeyer, W., Ellis, M. H. & Chandler, P. M. Semidwarf (sd-1), “green revolution” rice, contains a defective gibberellin 20-oxidase gene. Proc. Natl Acad. Sci. USA 99, 9043–9048 (2002).
Fan, L. et al. Post-domestication selection in the maize starch pathway. PLoS ONE 4, e7612 (2009).
Stumpf, M. P. & McVean, G. A. Estimating recombination rates from population-genetic data. Nature Rev. Genet. 4, 959–968 (2003).
Paterson, A. H. et al. The Sorghum bicolour genome and the diversification of grasses. Nature 457, 551–556 (2009).
McMullen, M. D. et al. Genetic properties of the maize nested association mapping population. Science 325, 737–740 (2009).
Schon, C. C., Dhillon, B. S., Utz, H. F. & Melchinger, A. E. High congruency of QTL positions for heterosis of grain yield in three crosses of maize. Theor. Appl. Genet. 120, 321–332 (2010).
Allard, R. W. History of plant population genetics. Annu. Rev. Genet. 33, 1–27 (1999).
Nordborg, M. Linkage disequilibrium, gene trees and selfing: an ancestral recombination graph with partial self-fertilization. Genetics 154, 923–929 (2000).
Morrell, P. L., Toleno, D. M., Lundy, K. E. & Clegg, M. T. Low levels of linkage disequilibrium in wild barley (Hordeum vulgare ssp. spontaneum) despite high rates of self-fertilization. Proc. Natl Acad. Sci. USA 102, 2442–2447 (2005).
Morrell, P. L., Toleno, D. M., Lundy, K. E. & Clegg, M. T. Estimating the contribution of mutation, recombination and gene conversion in the generation of haplotypic diversity. Genetics 173, 1705–1723 (2006).
Charlesworth, D. Effects of inbreeding on the genetic diversity of populations. Phil. Trans. R. Soc. Lond. B 358, 1051–1070 (2003).
Zhao, K. Y. et al. An Arabidopsis example of association mapping in structured samples. PLoS Genet. 3, e4 (2007).
Turner, A., Beales, J., Faure, S., Dunford, R. P. & Laurie, D. A. The pseudo-response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science 310, 1031–1034 (2005).
Yano, M. et al. Identification of quantitative trait loci controlling heading date in rice using a high-density linkage map. Theor. Appl. Genet. 95, 1025–1032 (1997).
Lin, Y. R., Schertz, K. F. & Paterson, A. H. Comparative analysis of QTLs affecting plant height and maturity across the Poaceae, in reference to an interspecific Sorghum population. Genetics 141, 391–411 (1995).
Gan, X. et al. Multiple reference genomes and transcriptomes for Arabidopsis thaliana. Nature 477, 419–423 (2011). De novo sequencing and annotation along with trascriptome sequencing of 18 reference genomes from the founders of a next-generation population are discussed in this paper. Re-annotation of individual genes suggests that many genes that appear to have lost function in simple comparisons with the original A. thaliana reference genome contain compensatory mutations that restore function at the locus.
Muller, H. J. Our load of mutations. Am. J. Hum. Genet. 2, 111–176 (1950).
Ng, S. B. et al. Exome sequencing identifies the cause of a Mendelian disorder. Nature Genet. 42, 30–35 (2010).
Gossmann, T. I. et al. Genome wide analyses reveal little evidence for adaptive evolution in many plant species. Mol. Biol. Evol. 27, 1822–1832 (2010).
Lai, J. S. et al. Genome-wide patterns of genetic variation among elite maize inbred lines. Nature Genet. 42, 1027–1030 (2010).
Gunther, T. & Schmid, K. J. Deleterious amino acid polymorphisms in Arabidopsis thaliana and rice. Theor. Appl. Genet. 121, 157–168 (2010).
Lu, J. et al. The accumulation of deleterious mutations in rice genomes: a hypothesis on the cost of domestication. Trends Genet. 22, 126–131 (2006).
Tang, H. B., Sezen, U. & Paterson, A. H. Domestication and plant genomes. Curr. Opin. Plant Biol. 13, 160–166 (2010).
Chun, S. & Fay, J. C. Evidence for hitchhiking of deleterious mutations within the human genome. PLoS Genet. 7, e1002240 (2011).
Lande, R. & Schemske, D. W. The evolution of self-fertilization and inbreeding depression in plants. I. Genetic models. Evolution 39, 24–40 (1985).
Charlesworth, D. & Willis, J. H. The genetics of inbreeding depression. Nature Rev. Genet. 10, 783–796 (2009).
Felsenstein, J. The evolutionary advantage of recombination. Genetics 78, 737–756 (1974).
Lynch, M., Conery, J. & Burger, R. Mutational meltdowns in sexual populations. Evolution 49, 1067–1080 (1995).
Meuwissen, T. H., Hayes, B. J. & Goddard, M. E. Prediction of total genetic value using genome-wide dense marker maps. Genetics 157, 1819–1829 (2001).
Heffner, E. L., Sorrells, M. E. & Jannink, J. L. Genomic selection for crop improvement. Crop Sci. 49, 1–12 (2009).
Andolfatto, P. et al. Multiplexed shotgun genotyping for rapid and efficient genetic mapping. Genome Res. 21, 610–617 (2011).
Elshire, R. J. et al. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 6, e19379 (2011).
Heffner, E. L., Jannink, J.-L. & Sorrells, M. E. Genomic selection accuracy using multifamily prediction models in a wheat breeding program. Plant Gen. 4, 65–75 (2011).
Salome, P. A. et al. Genetic architecture of flowering-time variation in Arabidopsis thaliana. Genetics 188, 421–433 (2011).
Troyer, A. F. Adaptedness and heterosis in corn and mule hybrids. Crop Sci. 46, 528–543 (2006).
Duvick, D. N. The contribution of breeding to yield advances in maize (Zea mays L.). Adv. Agron. 86, 83–145 (2005).
Bernardo, R. Molecular markers and selection for complex traits in plants: Learning from the last 20 years. Crop Sci. 48, 1649–1664 (2008).
Weinthal, D., Tovkach, A., Zeevi, V. & Tzfira, T. Genome editing in plant cells by zinc finger nucleases. Trends Plant Sci. 15, 308–321 (2010).
Bogdanove, A. J. & Voytas, D. F. TAL effectors: customizable proteins for DNA targeting. Science 333, 1843–1846 (2011).
Shukla, V. K. et al. Precise genome modification in the crop species Zea mays using zinc-finger nucleases. Nature 459, 437–441 (2009). This paper is an impressive demonstration of the potential power of targeted genomic editing. The authors use a custom zinc finger nuclease to modify two traits in maize and show that the method is sufficiently precise to target only one of the two paralogues of the enzyme of interest.
Morbitzer, R., Romer, P., Boch, J. & Lahaye, T. Regulation of selected genome loci using de novo-engineered transcription activator-like effector (TALE)-type transcription factors. Proc. Natl Acad. Sci. USA 107, 21617–21622 (2010).
Century, K., Reuber, T. L. & Ratcliffe, O. J. Regulating the regulators: the future prospects for transcription-factor-based agricultural biotechnology products. Plant Physiol. 147, 20–29 (2008).
Presgraves, D. C. The molecular evolutionary basis of species formation. Nature Rev. Genet. 11, 175–180 (2010).
Gepts, P. A comparison between crop domestication, classical plant breeding, and genetic engineering. Crop Sci. 42, 1780–1790 (2002).
Ming, R. et al. The draft genome of the transgenic tropical fruit tree papaya (Carica papaya Linnaeus). Nature 452, 991–996 (2008).
Argout, X. et al. The genome of Theobroma cacao. Nature Genet. 43, 101–108 (2011).
Goff, S. A. et al. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296, 92–100 (2002).
Varshney, R. K. et al. Draft genome sequence of pigeonpea (Cajanus cajan), an orphan legume crop of resource-poor farmers. Nature Biotech. 6 Nov 2011 (doi:10.1038/nbt.2022).
Schmutz, J. et al. Genome sequence of the palaeopolyploid soybean. Nature 463, 178–183 (2010).
Schnable, P. S. et al. The B73 maize genome: complexity, diversity, and dynamics. Science 326, 1112–1115 (2009).
Acknowledgements
We thank the US Department of Agriculture (USDA)–US National Institute for Food and Agriculture (NIFA) (2011-68002-30029) for partial support for P.L.M., USDA–NIFA (2009-01864) for J.R.-I. and the USDA Agriculture Research Service and the National Science Foundation (NSF)–Plant Genome Research (0820691) and NSF–Division of Biological Infrastructure (0965342) for support to E.S.B. The authors are grateful to J. Gerke, A. Gonzales, M. Hufford, D. Segal, R. Stupar and three anonymous referees for comments on the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Related links
FURTHER INFORMATION
Jeffrey Ross-Ibarra's homepage
1000 Genomes Project (in humans)
1001 Genomes Project (in A. thaliana)
Drosophila Genetic Reference Panel
International Barley Sequencing Consortium
International Tomato Genome Sequencing Project
International Wheat Genome Sequencing Consortium
US Department of Agriculture (USDA) National Agricultural Statistics Service
Glossary
- Genome-wide association studies
-
(GWASs). Studies that search for a statistical association between a phenotype and a particular allele by screening loci (most commonly by genotyping SNPs) across the entire genome.
- Paralogy
-
Unlike orthologous genes, which trace their common origin to a locus in an ancestral species, paralogous loci consist of gene copies that trace their common origin to a duplication event within a genome.
- Linkage disequilibrium
-
(LD). Nonrandom association of alleles at two or more loci. The pattern and extent of LD in a genomic region is affected by mutation, recombination, genetic drift, natural selection and demographic history.
- Bottleneck
-
A temporary marked reduction in population size.
- Site frequency spectrum
-
The distribution of allele frequencies in a population: essentially a count of the number of alleles in a population at a given frequency.
- Genetic drift
-
Fluctuations in allele frequencies that are due to the effects of random sampling.
- Admixture
-
The mixing of two or more genetically differentiated populations.
- Introgression
-
The incorporation of genetic material from one population or species into another by hybridization and backcrossing.
- Haplotype
-
The combination of alleles or genetic markers found on a single chromosome of an individual.
- Selective sweeps
-
Increases in frequency of an allele and closely linked chromosomal segments that are due to positive selection. Sweeps initially reduce variation and subsequently lead to a local excess of rare alleles as new unique mutations accumulate.
- Standing variation
-
Variation for a locus or trait that is polymorphic in a population.
- Heterosis
-
Otherwise known as 'hybrid vigour', heterosis is the phenomenon whereby progeny of a cross between genetically distinct parents have greater fitness than either of the parental types.
- Purifying selection
-
Selection against a deleterious allele.
- Ascertainment bias
-
Sampling bias that arises from how SNPs are chosen for inclusion on SNP arrays; SNPs that are known to be polymorphic in a particular population will have frequencies that are higher than would be expected by random sampling alone.
- Hill–Robertson effect
-
The reduction in efficacy of selection at a locus owing to selection at linked loci.
- Dobzhansky–Muller effects
-
Intrinsic reductions in viability or fertility resulting from epistatic interactions between multiple substitutions, typically observed in the offspring of a cross between individuals from genetically distinct populations.
Rights and permissions
About this article
Cite this article
Morrell, P., Buckler, E. & Ross-Ibarra, J. Crop genomics: advances and applications. Nat Rev Genet 13, 85–96 (2012). https://doi.org/10.1038/nrg3097
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrg3097
This article is cited by
-
Genetic insights into agronomic and morphological traits of drug-type cannabis revealed by genome-wide association studies
Scientific Reports (2024)
-
Epigenetics: Toward improving crop disease resistance and agronomic characteristics
Plant Biotechnology Reports (2024)
-
Physiological, biochemical, and molecular responses of rice (Oryza sativa L.) towards elevated ozone tolerance
Cereal Research Communications (2023)
-
Genome-wide identification and comparative evolutionary analysis of sorbitol metabolism pathway genes in four Rosaceae species and three model plants
BMC Plant Biology (2022)
-
COMPILE: a GWAS computational pipeline for gene discovery in complex genomes
BMC Plant Biology (2022)