Key Points
-
Sex chromosomes are unusually labile systems, with frequent shifts between male and female heterogamety and with a large variety in the precise number and types of chromosomes.
-
The lability is further emphasized by rapid turnover of genes and gene organization on the Y chromosome.
-
All types of sex chromosome (X, Y, Z and W) contain ampliconic structures of multi-copy genes.
-
A common model of sex-chromosome evolution implies gradual cessation of recombination between the proto-sex chromosomes.
-
The gene content of the X chromosome and the Z chromosome is unusual, with a non-random representation of genes with sex-biased gene expression.
-
The molecular evolution of sex-linked genes differs from autosomal genes with respect to mutation rate and selective pressure.
Abstract
It is now clear that sex chromosomes differ from autosomes in many aspects of genome biology, such as organization, gene content and gene expression. Moreover, sex linkage has numerous evolutionary genetic implications. Here, I provide a coherent overview of sex-chromosome evolution and function based on recent data. Heteromorphic sex chromosomes are almost as widespread across the animal and plant kingdoms as sexual reproduction itself and an accumulating body of genetic data reveals interesting similarities, as well as dissimilarities, between organisms with XY or ZW sex-determination systems. Therefore, I discuss how patterns and processes associated with sex linkage in male- and female-heterogametic systems offer a useful contrast in the study of sex-chromosome evolution.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Change history
16 September 2011
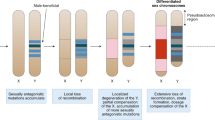
In figure 1a of the above article, the evolutionary strata (S1–S5) on the human X chromosome were incorrectly labelled. The figure showed the strata in the order S1 to S5, with S1 adjacent to the pseudoautosomal region. The order should be reversed (S5 to S1), with S5 adjacent to the pseudoautosomal region instead. The figure has been amended accordingly. The author apologizes for this error.
References
Maynard Smith, J. The Evolution of Sex (Cambridge Univ. Press, 1978).
Michod, R. E. & Levin, B. R. The Evolution of Sex: An Examination of Current Ideas (Sinauer Associates, Massachusetts, 1988).
Bull, J. J. Evolution of Sex Determining Mechanisms. (Benjamin-Cummings, California, 1981).
Charlesworth, B. The evolution of chromosomal sex determination. Novartis Found. Symp. 244, 207–219 (2002).
Gurbich, T. A. & Bachtrog, D. Gene content evolution on the X chromosome. Curr. Opin. Genet. Dev. 18, 493–498 (2008).
Vicoso, B. & Charlesworth, B. Evolution on the X chromosome: unusual patterns and processes. Nature Rev. Genet. 7, 645–653 (2006).
Presgraves, D. C. Sex chromosomes and speciation in Drosophila. Trends Genet. 24, 336–343 (2008).
Mank, J. E., Promislow, D. E. L. & Avise, J. C. Evolution of alternative sex-determining mechanisms in teleost fishes. Biol. J. Linn. Soc. 87, 83–93 (2006).
Hillis, D. M. & Green, D. M. Evolutionary changes of heterogametic sex in the phylogenetic history of amphibians. J. Evol. Biol. 3, 49–64 (1990).
Ogata, M., Hasegawa, Y., Ohtani, H., Mineyama, M. & Miura, I. The ZZ/ZW sex-determining mechanism originated twice and independently during evolution of the frog, Rana rugosa. Heredity 100, 92–99 (2007).
Fridolfsson, A. K. et al. Evolution of the avian sex chromosomes from an ancestral pair of autosomes. Proc. Natl Acad. Sci. USA 95, 8147–8152 (1998).
Takehana, Y., Hamaguchi, S. & Sakaizumi, M. Different origins of ZZ/ZW sex chromosomes in closely related medaka fishes, Oryzias javanicus. Chromosome Res. 16, 801–811 (2008).
Tanaka, K., Takehana, Y., Naruse, K., Hamaguchi, S. & Sakaizumi, M. Evidence for different origins of sex chromosomes in closely related Oryzias fishes: substitution of the master sex-determining gene. Genetics 177, 2075–2081 (2007).
van Doorn, G. S. & Kirkpatrick, M. Turnover of sex chromosomes induced by sexual conflict. Nature 449, 909–912 (2007).
van Doorn, G. S. & Kirkpatrick, M. Transitions between male and female heterogamety caused by sex-antagonistic selection. Genetics 186, 629–645 (2010).
Gruetzner, F., Ashley, T., Rowell, D. & Marshall Graves, J. How did the platypus get its sex chromosome chain? A comparison of meiotic multiples and sex chromosomes in plants and animals. Chromosoma 115, 75–88 (2006).
Charlesworth, D. & Mank, J. E. The birds and the bees and the flowers and the trees: lessons from genetic mapping of sex determination in plants and animals. Genetics 186, 9–31 (2010).
Marin, I. & Baker, B. S. The evolutionary dynamics of sex determination. Science 281, 1990–1994 (1998).
Raymond, C. S. et al. Evidence for evolutionary conservation of sex-determining genes. Nature 391, 691–695 (1998).
Raymond, C. S. et al. A region of human chromosome 9p required for testis development contains two genes related to known sexual regulators. Hum. Mol. Genet. 8, 989–996 (1999).
Smith, C. A. et al. The avian Z-linked gene DMRT1 is required for male sex determination in the chicken. Nature 461, 267–271 (2009).
Ellegren, H. et al. Faced with inequality: chicken do not have a general dosage compensation of sex-linked genes. BMC Biol. 5, 40 (2007).
Itoh, Y. et al. Dosage compensation is less effective in birds than in mammals. J. Biol. 6, 2 (2007).
Bergero, R. & Charlesworth, D. The evolution of restricted recombination in sex chromosomes. Trends Ecol. Evol. 24, 94–102 (2009).
Garcia-Moreno, J. & Mindell, D. P. Rooting a phylogeny with homologous genes on opposite sex chromosomes (gametologs): a case study using avian CHD. Mol. Biol. Evol. 17, 1826–1832 (2000).
Lahn, B. T. & Page, D. C. Four evolutionary strata on the human X chromosome. Science 286, 964–967 (1999).
Ross, M. T. et al. The DNA sequence of the human X chromosome. Nature 434, 325–337 (2005).
Nicolas, M. et al. A gradual process of recombination restriction in the evolutionary history of the sex chromosomes in dioecious plants. PLoS Biol. 3, e4 (2004).
Nam, K. & Ellegren, H. The chicken (Gallus gallus) Z chromosome contains at least three nonlinear evolutionary strata. Genetics 180, 1131–1136 (2008).
Menkis, A., Jacobson, D. J., Gustafsson, T. & Johannesson, H. The mating-type chromosome in the filamentous ascomycete Neurospora tetrasperma represents a model for early evolution of sex chromosomes. PLoS Genet. 4, e1000030 (2008).
Votintseva, A. A. & Filatov, D. A. Evolutionary strata in a small mating-type-specific region of the smut fungus Microbotryum violaceum. Genetics 182, 1391–1396 (2009).
Lemaitre, C. et al. Footprints of inversions at present and past pseudoautosomal boundaries in human sex chromosomes. Genome Biol. Evol. 1, 56–66 (2009).
Traut, W., Sahara, K. & Marec, F. Sex chromosomes and sex determination in Lepidoptera. Sexual Dev. 1, 332–346 (2007).
Veltsos, P., Keller, I. & Nichols, R. A. The inexorable spread of a newly arisen neo-Y chromosome. PLoS Genet. 4, e1000082 (2008).
Skaletsky, H. et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature 423, 825–837 (2003). By sequencing the human Y chromosome, this study revealed a hitherto unknown repetitive structure of multi-copy, testis-specific genes.
Repping, S. et al. High mutation rates have driven extensive structural polymorphism among human Y chromosomes. Nature Genet. 38, 463–467 (2006).
Rozen, S. et al. Abundant gene conversion between arms of palindromes in human and ape Y chromosomes. Nature 423, 873–876 (2003).
Bhowmick, B. K., Satta, Y. & Takahata, N. The origin and evolution of human ampliconic gene families and ampliconic structure. Genome Res. 17, 441–450 (2007).
Marais, G. A. B., Campos, P. R. A. & Gordo, I. Can intra-Y gene conversion oppose the degeneration of the human Y chromosome? A simulation study. Genome Biol. Evol. 2, 347–357 (2010).
Connallon, T. & Clark, A. G. Gene duplication, gene conversion, and the evolution of the Y chromosome. Genetics 186, 277–286 (2010).
Trombetta, B., Cruciani, F., Underhill, P. A., Sellitto, D. & Scozzari, R. Footprints of X-to-Y gene conversion in recent human evolution. Mol. Biol. Evol. 27, 714–725 (2010).
Rosser, Z. H., Balaresque, P. & Jobling, M. A. Gene conversion between the X chromosome and the male-specific region of the Y chromosome at a translocation hotspot. Am. J. Hum. Genet. 85, 130–134 (2009).
Hughes, J. F. et al. Chimpanzee and human Y chromosomes are remarkably divergent in structure and gene content. Nature 463, 536–539 (2010).
Koerich, L. B., Wang, X., Clark, A. G. & Carvalho, A. B. Low conservation of gene content in the Drosophila Y chromosome. Nature 456, 949–951 (2008).
Backström, N., Ceplitis, H., Berlin, S. & Ellegren, H. Gene conversion drives the evolution of HINTW, an ampliconic gene on the female-specific avian W chromosome. Mol. Biol. Evol. 22, 1992–1999 (2005).
Melamed, E. & Arnold, A. The role of LINEs and CpG islands in dosage compensation on the chicken Z chromosome. Chromosome Res. 17, 727–736 (2009).
Bailey, J. A., Carrel, L., Chakravarti, A. & Eichler, E. E. Molecular evidence for a relationship between LINE-1 elements and X chromosome inactivation: the Lyon repeat hypothesis. Proc. Natl Acad. Sci. USA 97, 6634–6639 (2000).
Bellott, D. W. et al. Convergent evolution of chicken Z and human X chromosomes by expansion and gene acquisition. Nature 466, 612–616 (2010). This study revealed unexpected similarities in gene organization between the human X and chicken Z chromosome.
Ellegren, H. & Parsch, J. The evolution of sex-biased genes and sex-biased gene expression. Nature Rev. Genet. 8, 689–698 (2007).
Innocenti, P. & Morrow, E. H. The sexually antagonistic genes of Drosophila melanogaster. PLoS Biol. 8, e1000335 (2010). The results of this study suggest that only a small proportion of genes with sex-biased expression are sexually antagonistic.
Rice, W. R. Sexually antagonistic genes: experimental evidence. Science 256, 1436–1439 (1992).
Rice, W. R. Sex chromosomes and the evolution of sexual dimorphism. Evolution 38, 735–742 (1984). Together with reference 99, this theoretical study represents a landmark that set the stage for expectations regarding DNA sequence evolution on sex chromosomes.
Sturgill, D., Zhang, Y., Parisi, M. & Oliver, B. Demasculinization of X chromosomes in the Drosophila genus. Nature 450, 238–241 (2007).
Parisi, M. et al. Paucity of genes on the Drosophila X chromosome showing male-biased expression. Science 299, 697–700 (2003). One of the first studies to use genome-wide gene expression data to demonstrate sex-biased gene expression.
Vicoso, B. & Charlesworth, B. The deficit of male-biased genes on the D. melanogaster X chromosome is expression-dependent: a consequence of dosage compensation? J. Mol. Evol. 68, 576–583 (2009).
Turner, J. M. A. Meiotic sex chromosome inactivation. Development 134, 1823–1831 (2007).
Wu, C. I. & Yujun Xu, E. Sexual antagonism and X inactivation — the SAXI hypothesis. Trends Genet. 19, 243–247 (2003).
Vibranovski, M. D., Lopes, H. F., Karr, T. L. & Long, M. Stage-specific expression profiling of Drosophila spermatogenesis suggests that meiotic sex chromosome inactivation drives genomic relocation of testis-expressed genes. PLoS Genet. 5, e1000731 (2009).
Parsch, J. X chromosome: expression and escape. PLoS Genet. 5, e1000724 (2009).
Hense, W., Baines, J. F. & Parsch, J. X chromosome inactivation during Drosophila spermatogenesis. PLoS Biol. 5, e273 (2007).
Vibranovski, M. D., Chalopin, D. S., Lopes, H. F., Long, M. & Karr, T. L. Direct evidence for postmeiotic transcription during Drosophila melanogaster spermatogenesis. Genetics 186, 431–433 (2010).
Khil, P. P., Smirnova, N. A., Romanienko, P. J. & Camerini-Otero, R. D. The mouse X chromosome is enriched for sex-biased genes not subject to selection by meiotic sex chromosome inactivation. Nature Genet. 36, 642–646 (2004).
Wang, P. J., McCarrey, J. R., Yang, F. & Page, D. C. An abundance of X-linked genes expressed in spermatogonia. Nature Genet. 27, 422–426 (2001).
Lercher, M. J., Urrutia, A. O. & Hurst, L. D. Evidence that the human X chromosome Is enriched for male-specific but not female-specific genes. Mol. Biol. Evol. 20, 1113–1116 (2003).
Zhang, Y. E., Vibranovski, M. D., Landback, P., Marais, G. A. B. & Long, M. Chromosomal redistribution of male-biased genes in mammalian evolution with two bursts of gene gain on the X chromosome. PLoS Biol. 8, e1000494 (2010).
Kaiser, V. B. & Ellegren, H. Nonrandom distribution of genes with sex-biased expression in the chicken genome. Evolution 60, 1945–1951 (2006).
Storchová, R. & Divina, P. Nonrandom representation of sex-biased genes on chicken Z chromosome. J. Mol. Evol. 63, 676–681 (2006).
Mank, J. E. & Ellegren, H. Sex-linkage of sexually antagonistic genes is predicted by female, but not male, effects in birds. Evolution 63, 1464–1472 (2009).
Morˇkovský, L. et al. The chicken Z chromosome is enriched for genes with preferential expression in ovarian somatic cells. J. Mol. Evol. 70, 129–136 (2009).
Schoenmakers, S. et al. Female meiotic sex chromosome inactivation in chicken. PLoS Genet. 5, e1000466 (2009).
Arunkumar, K. P., Mita, K. & Nagaraju, J. The silkworm Z chromosome is enriched in testis-specific genes. Genetics 182, 493–501 (2009).
Emerson, J. J., Kaessmann, H., Betran, E. & Long, M. Extensive gene traffic on the mammalian X chromosome. Science 303, 537–540 (2004).
Shiao, M. S. et al. Origins of new male germ-line functions from X-derived autosomal retrogenes in the mouse. Mol. Biol. Evol. 24, 2242–2253 (2007).
Vinckenbosch, N., Dupanloup, I. & Kaessmann, H. Evolutionary fate of retroposed gene copies in the human genome. Proc. Natl Acad. Sci. USA 103, 3220–3225 (2006).
Betran, E., Thornton, K. & Long, M. Retroposed new genes out of the X in Drosophila. Genome Res. 12, 1854–1859 (2002).
Meisel, R. P., Han, M. V. & Hahn, M. W. A complex suite of forces drives gene traffic from Drosophila X chromosomes. Genome Biol. Evol. 1, 176–188 (2009).
Vibranovski, M. D., Zhang, Y. & Long, M. General gene movement off the X chromosome in the Drosophila genus. Genome Res. 19, 897–903 (2009).
Zhang, Y. E., Vibranovski, M. D., Krinsky, B. H. & Long, M. Age-dependent chromosomal distribution of male-biased genes in Drosophila. Genome Res. 20, 1526–1533 (2010).
Pink, C. J. et al. Evidence that replication-associated mutation alone does not explain between-chromosome differences in substitution rates. Genome Biol. Evol. 1, 13–22 (2009).
Crow, J. F. The origins, patterns and implications of human spontaneous mutation. Nature Rev. Genet. 1, 40–47 (2000).
Li, W.-H., Yi, S. & Makova, K. Male-driven evolution. Curr. Opin. Genet. Dev. 12, 650–656 (2002).
Shimmin, L. C., Chang, B. H.-J. & Li, W.-H. Male-driven evolution of DNA sequences. Nature 362, 745–747 (1993).
Ellegren, H. & Fridolfsson, A.-K. Sex-specific mutation rates in salmonid fish. J. Mol. Evol. 56, 458–463 (2003). This study provided conclusive evidence for male-biased mutation, a concept that had been questioned because the observation in mammals of a higher rate of neutral divergence on the Y than on the X chromosome could be interpreted in terms of a specifically reduced X chromosome rate, unrelated to sex-specific mutation rates.
Bachtrog, D. Evidence for male-driven evolution in Drosophila. Mol. Biol. Evol. 25, 617–619 (2008).
Ellegren, H. & Fridolfsson, A.-K. Male-driven evolution of DNA sequences in birds. Nature Genet. 17, 182–184 (1997).
Charlesworth, B. & Charlesworth, D. The degeneration of Y chromosomes. Phil. Trans. R. Soc. B 355, 1563–1572 (2000). A seminal review on the evolutionary processes that affect non-recombining chromosomes.
Bachtrog, D. Evidence that positive selection drives Y-chromosome degeneration in Drosophila miranda. Nature Genet. 36, 518–522 (2004).
Kaiser, V. B. & Charlesworth, B. Muller's ratchet and the degeneration of the Drosophila miranda neo-Y chromosome. Genetics 185, 339–348 (2010).
Kaiser, V. B. & Charlesworth, B. The effects of deleterious mutations on evolution in non-recombining genomes. Trends Genet. 25, 9–12 (2009).
Bachtrog, D. The temporal dynamics of processes underlying Y chromosome degeneration. Genetics 179, 1513–1525 (2008).
Engelstadter, J. Muller's ratchet and the degeneration of Y chromosomes: a simulation study. Genetics 180, 957–967 (2008).
International Chicken Genome Sequencing Consortium. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 432, 695–716 (2004).
Berlin, S. & Ellegren, H. Fast accumulation of nonsynonymous mutations on the female-specific W chromosome in birds. J. Mol. Evol. 62, 66–72 (2006).
Bachtrog, D. & Charlesworth, B. Reduced levels of microsatellite variability on the neo-Y chromosome of Drosophila miranda. Curr. Biol. 10, 1025–1031 (2000).
Filatov, D. A., Moneger, F., Negrutiu, I. & Charlesworth, D. Low variability in a Y-linked plant gene and its implications for Y-chromosome evolution. Nature 404, 388–390 (2000). The results of this study provided evidence that similar processes are associated with sex-chromosome evolution in plants and animals.
Hellborg, L. & Ellegren, H. Low levels of nucleotide diversity in mammalian Y chromosomes. Mol. Biol. Evol. 21, 158–163 (2004).
Rozen, S., Marszalek, J. D., Alagappan, R. K., Skaletsky, H. & Page, D. C. Remarkably little variation in proteins encoded by the Y chromosome's single-copy genes, implying effective purifying selection. Am. J. Hum. Genet. 85, 923–928 (2009).
The International SNP Map Working Group. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature 409, 928–933 (2001).
Berlin, S. & Ellegren, H. Chicken W: a genetically uniform chromosome in a highly variable genome. Proc. Natl Acad. Sci. USA 101, 15967–15969 (2004).
Charlesworth, B., Coyne, J. A. & Barton, N. H. The relative rates of evolution of sex chromosomes and autosomes. Am. Nat. 130, 113 (1987).
Mank, J. E., Vicoso, B., Berlin, S. & Charlesworth, B. Effective population size and the faster-X effect: empirical results and their interpretation. Evolution 64, 663–674 (2010).
Mank, J. E., Axelsson, E. & Ellegren, H. Fast-X on the Z: rapid evolution of sex-linked genes in birds. Genome Res. 17, 618–624 (2007).
Mank, J. E., Nam, K. & Ellegren, H. Faster-Z evolution is predominantly due to genetic drift. Mol. Biol. Evol. 27, 661–670 (2009).
Ellegren, H. The different levels of genetic diversity in sex chromosomes and autosomes. Trends Genet. 25, 278–284 (2009).
Bustamante, C. D. & Ramachandran, S. Evaluating signatures of sex-specific processes in the human genome. Nature Genet. 41, 8–10 (2009).
Pool, J. E. & Nielsen, R. Population size changes reshape genomic patterns of diversity. Evolution 61, 3001–3006 (2007).
Hammer, M. F. et al. The ratio of human X chromosome to autosome diversity is positively correlated with genetic distance from genes. Nature Genet. 42, 803–831 (2010). This paper demonstrates that chromosome-specific estimates of genetic diversity are dependent on the genetic distance between marker loci and potential targets of selection.
Charlesworth, D. Plant sex determination and sex chromosomes. Heredity 88, 94–101 (2002).
Shetty, S., Griffin, D. K. & Graves, J. A. M. Comparative painting reveals strong chromosome homology over 80 million years of bird evolution. Chromosome Res. 7, 289–295 (1999).
Kawai, A. et al. The ZW sex chromosomes of Gekko hokouensis (Gekkonidae, Squamata) represent highly conserved homology with those of avian species. Chromosoma 118, 43–51 (2009).
Matsubara, K. et al. Evidence for different origin of sex chromosomes in snakes, birds, and mammals and step-wise differentiation of snake sex chromosomes. Proc. Natl Acad. Sci. USA 103, 18190–18195 (2006).
Coyne, J. A. Genetics and speciation. Nature 355, 511–515 (1992).
Coyne, J. A. & Orr, H. A. in Speciation and its Consequences (eds Otte, D. & Endler, J.) 180–207 (Sinaur Associates, 1989).
Masly, J. P. & Presgraves, D. C. High-resolution genome-wide dissection of the two rules of speciation in Drosophila. PLoS Biol. 5, e243 (2007).
Lifschytz, E. & Lindsley, D. L. The role of X-chromosome inactivation during spermatogenesis. Proc. Natl Acad. Sci. USA 69, 182–186 (1972).
Good, J. M., Giger, T., Dean, M. D. & Nachman, M. W. Widespread over-expression of the X chromosome in sterile F1 hybrid mice. PLoS Genet. 6, e1001148 (2010).
Lu, X. et al. Genome-wide misexpression of X-linked versus autosomal genes associated with hybrid male sterility. Genome Res. 20, 1097–1102 (2010).
Waters, P. D., Duffy, B., Frost, C. J., Delbridge, M. L. & Graves, J. A. M. The human Y chromosome derives largely from a single autosomal region added to the sex chromosomes 80–130 million years ago. Cytogenet. Cell Genet. 92, 74–79 (2001).
Page, D. C., Harper, M. E., Love, J. & Botstein, D. Occurrence of a transposition from the X-chromosome long arm to the Y-chromosome short arm during human evolution. Nature 311, 119–123 (1984).
Acknowledgements
Work in my laboratory is supported by the Swedish Research Council, a European Research Council Advanced Investigator Grant and a Knut and Alice Wallenberg Foundation Wallenberg Scholar Award. I thank S. Adolfsson and H. Johannesson for useful comments on the manuscript.
Author information
Authors and Affiliations
Ethics declarations
Competing interests
The author declares no competing financial interests.
Related links
Glossary
- Sexual antagonism
-
Sexually antagonistic genes are genes for which expression has contrasting effects on fitness in the two sexes.
- Gametologous
-
Gametologous genes are homologous genes shared between the X and Y, or Z and W, chromosomes that have evolved independently since recombination ceased in the ancestral gene.
- Pseudoautosomal region
-
(PAR). The region of both sex chromosomes that still recombines in the heterogametic sex. In old and highly differentiated sex chromosomes, such as the mammalian X and Y, the PAR is usually small.
- Gene conversion
-
The transfer of genetic material from one chromosomal region to another. The 'donor' locus remains intact whereas the 'acceptor' locus changes. Gene conversion occurs between more or less homologous sequences.
- Granulosa cells
-
Somatic cells in the ovary that surround the oocyte.
- Haldane's rule
-
The tendency for offspring of the heterogametic sex to suffer more severe fitness consequences from an interspecific mating (hybridization) than the homogametic sex.
- Effective population size
-
(Ne). Indicates how many individuals actually contribute alleles to the next generation, as opposed to the actual number of individuals in a population. For various reasons, including the preferential reproduction of some individuals and population size fluctuations over time, the effective population size is typically smaller than the actual number of individuals in the population.
- Muller's ratchet
-
A population of non-recombining chromosomes may — by chance or by selection for beneficial variants contained within other haplotypes — irreversibly lose the class of chromosomes that is least loaded with deleterious mutations. Like a ratchet, this is a unidirectional process that inevitably leads to the degeneration of non-recombining chromosomes.
- Hill–Robertson interference
-
The general concept of selection at one locus affecting the efficiency of selection at a linked locus.
- Selective sweep
-
A positive selection for an advantageous allele will increase the frequency of not only that allele but also other alleles contained within the same haplotype, causing a selective sweep (also referred to as genetic hitch-hiking). A hallmark of such sweeps is reduced levels of genetic diversity around the selected locus.
- Background selection
-
Purifying (negative) selection against a deleterious allele will also tend to remove linked variants, or at least decrease their frequency, contributing to loss of genetic diversity (compare with selective sweep).
- Chromosome painting
-
The use of an isolated chromosome, labelled with a fluorophore, as a probe in hybridization to a chromosome spread of the same or of a different species. The chromosomal regions homologous to the probe will be 'painted' and light up when fluorescence is detected.
Rights and permissions
About this article
Cite this article
Ellegren, H. Sex-chromosome evolution: recent progress and the influence of male and female heterogamety. Nat Rev Genet 12, 157–166 (2011). https://doi.org/10.1038/nrg2948
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrg2948
This article is cited by
-
Genetic sex test for the short-beaked echidna (Tachyglossus aculeatus)
Conservation Genetics Resources (2022)
-
Molecular evolution and the decline of purifying selection with age
Nature Communications (2021)
-
From molecules to populations: appreciating and estimating recombination rate variation
Nature Reviews Genetics (2020)
-
Sex chromosome evolution in parasitic nematodes of humans
Nature Communications (2020)