Key Points
-
Plant immunity depends on cell-autonomous events that are related to animal innate immunity, but plants have a greatly expanded recognition repertoire to compensate for their lack of an adaptive immune system. Ongoing research is revealing the recognition capacity of the plant immune system and concurrent studies on pathogen biology are beginning to unravel how these organisms manipulate host immunity to cause disease.
-
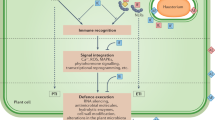
Plants have evolved two strategies to detect pathogens. On the external face of the host cell, conserved microbial elicitors called pathogen-associated molecular patterns (PAMPs) are recognized by receptor proteins called pattern recognition receptors (PRRs); stimulation of PRRs leads to PAMP-triggered immunity (PTI). The second class of perception involves recognition by intracellular receptors of pathogen virulence molecules called effectors; this recognition induces effector-triggered immunity (ETI).
-
PTI is generally effective against non-adapted pathogens in a phenomenon called non-host resistance, whereas ETI is active against adapted pathogens. However, these relationships are not exclusive and depend on the elicitor molecules present in each infection.
-
Successful pathogens are able to suppress PTI responses and thereby multiply and cause disease. They achieve suppression through the deployment of 'effector' proteins. Plant receptor proteins can recognize pathogen effectors either by direct physical association or indirectly through an accessory protein.
-
Our understanding of effector proteins and their host targets is at an early stage. Sophisticated biochemical screens for host protein targets that interact with the diverse suites of pathogen effectors is likely to lead to the identification of important components of host defence mechanisms, and teach us more about host immune pathways and pathogenicity strategies.
-
It is crucially important for the deployment of existing and novel resistance genes in agriculture that we advance our knowledge of plant–pathogen molecular co-evolution.
Abstract
Plants are engaged in a continuous co-evolutionary struggle for dominance with their pathogens. The outcomes of these interactions are of particular importance to human activities, as they can have dramatic effects on agricultural systems. The recent convergence of molecular studies of plant immunity and pathogen infection strategies is revealing an integrated picture of the plant–pathogen interaction from the perspective of both organisms. Plants have an amazing capacity to recognize pathogens through strategies involving both conserved and variable pathogen elicitors, and pathogens manipulate the defence response through secretion of virulence effector molecules. These insights suggest novel biotechnological approaches to crop protection.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Singh, R. P. et al. in Advances in Agronomy (ed. Sparks, D. L.) Vol. 98 Ch. 5, 271–309 (Elsevier, London, 2008).
Flor, H. H. Current status of gene-for-gene concept. Annu. Rev. Phytopathol. 9, 275–296 (1971).
Ausubel, F. M. Are innate immune signaling pathways in plants and animals conserved? Nature Immunol. 6, 973–979 (2005).
Chisholm, S. T., Coaker, G., Day, B. & Staskawicz, B. J. Host–microbe interactions: shaping the evolution of the plant immune response. Cell 124, 803–814 (2006).
Jones, J. D. & Dangl, J. L. The plant immune system. Nature 444, 323–329 (2006).
Boller, T. & Felix, G. A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60, 379–406 (2009).
Zipfel, C. Pattern-recognition receptors in plant innate immunity. Curr. Opin. Immunol. 20, 10–16 (2008).
Lehti-Shiu, M. D., Zou, C., Hanada, K. & Shiu, S.-H. Evolutionary history and stress regulation of plant receptor-like kinase/pelle genes. Plant Physiol. 150, 12–26 (2009).
Wang, G. et al. A genome-wide functional investigation into the roles of receptor-like proteins in Arabidopsis. Plant Physiol. 147, 503–517 (2008).
Gay, N. J. & Gangloff, M. Structure and function of Toll receptors and their ligands. Annu. Rev. Biochem. 76, 141–165 (2007).
Zipfel, C. et al. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125, 749–760 (2006).
Lee, S.-W. et al. A type I-secreted, sulfated peptide triggers XA21-mediated innate immunity. Science 326, 850–853 (2009). Although most PRRs provide subtle protection, this paper shows that a single PAMP receptor can confer effective disease resistance to a bacterial pathogen.
Chinchilla, D. et al. A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448, 497–500 (2007).
Heese, A. et al. The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc. Natl Acad. Sci. USA 104, 12217–12222 (2007). This study identified of BAK1 as a central regulator of plant immunity.
Miya, A. et al. CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl Acad. Sci. USA 104, 19613–19618 (2007).
Wan, J. et al. A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell 20, 471–481 (2008).
Gimenez-Ibanez, S., Ntoukakis, V. & Rathjen, J. The LysM receptor kinase CERK1 mediates bacterial perception in Arabidopsis. Plant Signal. Behav. 4, 539–541 (2009).
Schulze, B. et al. Rapid heteromerization and phosphorylation of ligand-activated plant transmembrane receptors and their associated kinase BAK1. J. Biol. Chem. 285, 9444–9451 (2010).
Shan, L. et al. Bacterial effectors target the common signaling partner BAK1 to disrupt multiple MAMP receptor-signaling complexes and impede plant immunity. Cell Host Microbe 4, 17–27 (2008).
Kemmerling, B. et al. The BRI1-associated kinase 1, BAK1, has a brassinolide-independent role in plant cell-death control. Curr. Biol. 17, 1116–1122 (2007).
He, K. et al. BAK1 and BKK1 regulate brassinosteroid-dependent growth and brassinosteroid-independent cell-death pathways. Curr. Biol. 17, 1109–1115 (2007).
Veronese, P. et al. The membrane-anchored BOTRYTIS-INDUCED KINASE1 plays distinct roles in Arabidopsis resistance to necrotrophic and biotrophic pathogens. Plant Cell 18, 257–273 (2006).
Lu, D. et al. A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc. Natl Acad. Sci. USA 107, 496–501 (2010).
Cunnac, S., Lindeberg, M. & Collmer, A. Pseudomonas syringae type III secretion system effectors: repertoires in search of functions. Curr. Opin. Microbiol. 12, 53–60 (2009).
Kvitko, B. H. et al. Deletions in the repertoire of Pseudomonas syringae pv. tomato DC3000 type III secretion effector genes reveal functional overlap among effectors. PLoS Pathog. 5, e1000388 (2009). A clear, genetic demonstration that bacterial effectors work redundantly. This explains why individual deletions of effector genes often have minor phenotypes.
Zhou, J.-M. & Chai, J. Plant pathogenic bacterial type III effectors subdue host responses. Curr. Opin. Microbiol. 11, 179–185 (2008).
Hauck, P., Thilmony, R. & He, S. Y. A Pseudomonas syringae type III effector suppresses cell wall-based extracellular defense in susceptible Arabidopsis plants. Proc. Natl Acad. Sci. USA 100, 8577–8582 (2003).
Kim, M. G. et al. Two Pseudomonas syringae type III effectors inhibit RIN4-regulated basal defense in Arabidopsis. Cell 121, 749–759 (2005).
Grant, S. R., Fisher, E. J., Chang, J. H., Mole, B. M. & Dangl, J. L. Subterfuge and manipulation: type III effector proteins of phytopathogenic bacteria. Annu. Rev. Microbiol. 60, 425–449 (2006).
Xiang, T. et al. Pseudomonas syringae effector AvrPto blocks innate immunity by targeting receptor kinases. Curr. Biol. 18, 74–80 (2008). This paper shows that the bacterial effector AvrPto targets receptor kinases, providing the intellectual basis for current decoy models of indirect pathogen recognition.
Gohre, V. et al. Plant pattern-recognition receptor FLS2 is directed for degradation by the bacterial ubiquitin ligase AvrPtoB. Curr. Biol. 18, 1824–1832 (2008).
Gimenez-Ibanez, S. et al. AvrPtoB targets the LysM receptor kinase CERK1 to promote bacterial virulence on plants. Curr. Biol. 19, 423–429 (2009).
Rosebrock, T. R. et al. A bacterial E3 ubiquitin ligase targets a host protein kinase to disrupt plant immunity. Nature 448, 370–374 (2007).
Xing, W. et al. The structural basis for activation of plant immunity by bacterial effector protein AvrPto. Nature 449, 243–247 (2007).
Axtell, M. J. & Staskawicz, B. J. Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell 112, 369–377 (2003).
Mackey, D., Belkhadir, Y., Alonso, J. M., Ecker, J. R. & Dangl, J. Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell 112, 379–389 (2003).
Wilton, M. et al. The type III effector HopF2Pto targets Arabidopsis RIN4 protein to promote Pseudomonas syringae virulence. Proc. Natl Acad. Sci. USA 107, 2349–2354 (2010).
Marathe, R. & Dinesh-Kumar, S. P. Plant defense: one post, multiple guards?! Mol. Cell 11, 284–286 (2003).
Liu, J. et al. RIN4 functions with plasma membrane H+-ATPases to regulate stomatal apertures during pathogen attack. PLoS Biol. 7, e1000139 (2009).
Kay, S. & Bonas, U. How Xanthomonas type III effectors manipulate the host plant. Curr. Opin. Microbiol. 12, 37–43 (2009).
Kay, S., Hahn, S., Marois, E., Hause, G. & Bonas, U. A bacterial effector acts as a plant transcription factor and Induces a cell size regulator. Science 318, 648–651 (2007).
Boch, J. et al. Breaking the code of DNA binding specificity of TAL-Type III effectors. Science 326, 1509–1512 (2009).
Moscou, M. J. & Bogdanove, A. J. A simple cipher governs DNA recognition by TAL effectors. Science 326, 1501 (2009).
Römer, P., Recht, S. & Lahaye, T. A single plant resistance gene promoter engineered to recognize multiple TAL effectors from disparate pathogens. Proc. Natl Acad. Sci. USA 106, 20526–20531 (2009).
Gu, K. et al. R. gene expression induced by a type-III effector triggers disease resistance in rice. Nature 435, 1122–1125 (2005).
Romer, P. et al. Plant pathogen recognition mediated by promoter activation of the pepper Bs3 resistance gene. Science 318, 645–648 (2007).
Kamoun, S. Groovy times: filamentous pathogen effectors revealed. Curr. Opin. Plant Biol. 10, 358–365 (2007).
Panstruga, R. & Dodds, P. N. Terrific protein traffic: the mystery of effector protein delivery by filamentous plant pathogens. Science 324, 748–750 (2009).
Haas, B. J. et al. Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature 461, 393–398 (2009).
Dean, R. A. et al. The genome sequence of the rice blast fungus Magnaporthe grisea. Nature 434, 980–986 (2005).
Kämper, J. et al. Insights from the genome of the biotrophic fungal plant pathogen Ustilago maydis. Nature 444, 97–101 (2006).
Hogenhout, S. A., Van der Hoorn, R. A. L., Terauchi, R. & Kamoun, S. Emerging concepts in effector biology of plant-associated organisms. Mol. Plant Microbe Interact. 22, 115–122 (2009).
Bos, J. I. et al. Phytophthora infestans effector AVR3a is essential for virulence and manipulates plant immunity by stabilizing host E3 ligase CMPG1. Proc. Natl Acad. Sci. USA 107, 9909–9914 (2010).
Bellafiore, S. et al. Direct identification of the Meloidogyne incognita secretome reveals proteins with host cell reprogramming potential. PLoS Pathog. 4, e1000192 (2008).
Voinnet, O. RNA silencing as a plant immune system against viruses. Trends Genet. 17, 449–459 (2001).
Girardin, S. E., Philpott, D. J. & Lemaitre, B. Sensing microbes by diverse hosts. Workshop on pattern recognition proteins and receptors. EMBO Rep. 4, 932–936 (2003).
Jia, Y., McAdams, S. A., Bryan, G. T., Hershey, H. P. & Valent, B. Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J. 19, 4004–4014 (2000).
Catanzariti, A. M., Dodds, P. N., Lawrence, G. J., Ayliffe, M. A. & Ellis, J. G. Haustorially expressed secreted proteins from flax rust are highly enriched for avirulence elicitors. Plant Cell 18, 243–256 (2006).
Dodds, P. N., Lawrence, G. J., Catanzariti, A. M., Ayliffe, M. A. & Ellis, J. G. The Melampsora lini AvrL567 avirulence genes are expressed in haustoria and their products are recognized inside plant cells. Plant Cell 16, 755–768 (2004).
Catanzariti, A. M. et al. The AvrM effector from flax rust has a structured C-terminal domain and interacts directly with the M resistance protein. Mol. Plant Microbe Interact. 23, 49–57 (2010).
Dodds, P. N. et al. Direct protein interaction underlies gene-for-gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes. Proc. Natl Acad. Sci. USA 103, 8888–8893 (2006). An elegant demonstration of how direct recognition of pathogen effectors works at the molecular level and drives antagonistic co-evolution.
van der Hoorn, R. A. & Kamoun, S. From guard to decoy: a new model for perception of plant pathogen effectors. Plant Cell 20, 2009–2017 (2008).
Dangl, J. L. & Jones, J. D. Plant pathogens and integrated defence responses to infection. Nature 411, 826–833 (2001).
Mackey, D., Holt, B. F., Wiig, A. & Dangl, J. L. RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell 108, 743–754 (2002).
Mucyn, T. S. et al. The tomato NBARC-LRR protein Prf interacts with Pto kinase in vivo to regulate specific plant immunity. Plant Cell 18, 2792–2806 (2006).
Zipfel, C. & Rathjen, J. P. Plant immunity: AvrPto targets the frontline. Curr. Biol. 18, R218–R220 (2008).
Gutierrez, J. R. et al. Prf immune complexes of tomato are oligomeric and contain multiple Pto-like kinases that diversify effector recognition. Plant J. 61, 507–581 (2009).
Mucyn, T. S., Wu, A. J., Balmuth, A. L., Arasteh, J. M. & Rathjen, J. P. Regulation of tomato Prf by Pto-like protein kinases. Mol. Plant Microbe Interact. 22, 391–401 (2009).
Collier, S. M. & Moffett, P. NB-LRRs work a 'bait and switch' on pathogens. Trends Plant Sci. 14, 521–529 (2009).
Ntoukakis, V. et al. Host inhibition of a bacterial virulence effector triggers immunity to infection. Science 324, 784–787 (2009).
Narusaka, M. et al. RRS1 and RPS4 provide a dual resistance-gene system against fungal and bacterial pathogens. Plant J. 60, 218–226 (2009). An as-yet-unique example showing how the host can inactivate a pathogen effector, leading to its recognition and host immunity.
Sinapidou, E. et al. Two TIR:NB:LRR genes are required to specify resistance to Peronospora parasitica isolate Cala2 in Arabidopsis. Plant J. 38, 898–909 (2004).
Loutre, C. et al. Two different CC-NBS-LRR genes are required for Lr10-mediated leaf rust resistance in tetraploid and hexaploid wheat. Plant J. 60, 1043–1054 (2009).
Lee, S. K. et al. Rice Pi5-mediated resistance to Magnaporthe oryzae requires the presence of two coiled-coil-nucleotide-binding-leucine-rich repeat genes. Genetics 181, 1627–1638 (2009).
Takken, F. L. & Tameling, W. I. To nibble at plant resistance proteins. Science 324, 744–746 (2009).
Birker, D. et al. A locus conferring resistance to Colletotrichum higginsianum is shared by four geographically distinct Arabidopsis accessions. Plant J. 60, 602–613 (2009).
Dodds, P. N., Lawrence, G. J. & Ellis, J. G. Six amino acid changes confined to the leucine-rich repeat β-strand/β-turn motif determine the difference between the P and P2 rust resistance specificities in flax. Plant Cell 13, 163–178 (2001).
Ellis, J. G., Lawrence, G. J., Luck, J. E. & Dodds, P. N. Identification of regions in alleles of the flax rust resistance gene L that determine differences in gene-for-gene specificity. Plant Cell 11, 495–506 (1999).
Shen, Q. H. et al. Recognition specificity and RAR1/SGT1 dependence in barley Mla disease resistance genes to the powdery mildew fungus. Plant Cell 15, 732–744 (2003).
Rairdan, G. J. & Moffett, P. Distinct domains in the ARC region of the potato resistance protein Rx mediate LRR binding and inhibition of activation. Plant Cell 18, 2082–2093 (2006).
Belkhadir, Y., Nimchuk, Z., Hubert, D. A., Mackey, D. & Dangl, J. L. Arabidopsis RIN4 negatively regulates disease resistance mediated by RPS2 and RPM1 downstream or independent of the NDR1 signal modulator and is not required for the virulence functions of bacterial type III effectors AvrRpt2 or AvrRpm1. Plant Cell 16, 2822–2835 (2004).
Tameling, W. I. et al. Mutations in the NB-ARC domain of I-2 that impair ATP hydrolysis cause autoactivation. Plant Physiol. 140, 1233–1245 (2006).
Inohara, N. et al. An induced proximity model for NF-κB activation in the Nod1/RICK and RIP signaling pathways. J. Biol. Chem. 275, 27823–27831 (2000).
Shaw, M. H., Reimer, T., Kim, Y. G. & Nunez, G. NOD-like receptors (NLRs): bona fide intracellular microbial sensors. Curr. Opin. Immunol. 20, 377–382 (2008).
Mestre, P. & Baulcombe, D. C. Elicitor-mediated oligomerization of the tobacco N disease resistance protein. Plant Cell 18, 491–501 (2006).
Frost, D. et al. Tobacco transgenic for the flax rust resistance gene L expresses allele-specific activation of defense responses. Mol. Plant Microbe Interact. 17, 224–232 (2004).
Swiderski, M. R., Birker, D. & Jones, J. D. The TIR domain of TIR-NB-LRR resistance proteins is a signaling domain involved in cell death induction. Mol. Plant Microbe Interact. 22, 157–165 (2009).
Bendahmane, A., Farnham, G., Moffett, P. & Baulcombe, D. C. Constitutive gain-of-function mutants in a nucleotide binding site-leucine rich repeat protein encoded at the Rx locus of potato. Plant J. 32, 195–204 (2002).
Rairdan, G. J. et al. The coiled-coil and nucleotide binding domains of the potato Rx disease resistance protein function in pathogen recognition and signaling. Plant Cell 20, 739–751 (2008).
Tao, Y., Yuan, F., Leister, R. T., Ausubel, F. M. & Katagiri, F. Mutational analysis of the Arabidopsis nucleotide binding site-leucine-rich repeat resistance gene RPS2. Plant Cell 12, 2541–2554 (2000).
Tao, Y. et al. Quantitative nature of Arabidopsis responses during compatible and incompatible interactions with the bacterial pathogen Pseudomonas syringae. Plant Cell 15, 317–330 (2003).
Pitzschke, A., Schikora, A. & Hirt, H. MAPK cascade signalling networks in plant defence. Curr. Opin. Plant Biol. 12, 421–426 (2009).
Asai, T. et al. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415, 977–983 (2002).
Suarez-Rodriguez, M. C. et al. MEKK1 is required for flg22-induced MPK4 activation in Arabidopsis plants. Plant Physiol. 143, 661–669 (2007).
Liu, Y. & Zhang, S. Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. Plant Cell 16, 3386–3399 (2004).
Bethke, G. et al. Flg22 regulates the release of an ethylene response factor substrate from MAP kinase 6 in Arabidopsis thaliana via ethylene signaling. Proc. Natl Acad. Sci. USA 106, 8067–8072 (2009).
Boudsocq, M. et al. Differential innate immune signalling via Ca2+ sensor protein kinases. Nature 464, 418–422 (2010).
Shirasu, K. & Schulze-Lefert, P. Complex formation, promiscuity and multi-functionality: protein interactions in disease-resistance pathways. Trends Plant Sci. 8, 252–258 (2003).
Wiermer, M., Feys, B. J. & Parker, J. E. Plant immunity: the EDS1 regulatory node. Curr. Opin. Plant Biol. 8, 383–389 (2005).
Day, B., Dahlbeck, D. & Staskawicz, B. J. NDR1 interaction with RIN4 mediates the differential activation of multiple disease resistance pathways in Arabidopsis. Plant Cell 18, 2782–2791 (2006).
Burch-Smith, T. M. et al. A novel role for the TIR domain in association with pathogen-derived elicitors. PLoS Biol. 5, e68 (2007).
Shen, Q. H. et al. Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science 315, 1098–1103 (2007).
Wirthmueller, L., Zhang, Y., Jones, J. D. & Parker, J. E. Nuclear accumulation of the Arabidopsis immune receptor RPS4 is necessary for triggering EDS1-dependent defense. Curr. Biol. 17, 2023–2029 (2007). The key paper underlying the hypothesis that an active fraction of plant NB-LRR proteins resides in the plant cell nucleus.
Deslandes, L. et al. Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc. Natl Acad. Sci. USA 100, 8024–8029 (2003).
Bernoux, M. et al. RD19, an Arabidopsis cysteine protease required for RRS1-R-mediated resistance, is relocalized to the nucleus by the Ralstonia solanacearum PopP2 effector. Plant Cell 20, 2252–2264 (2008).
Bari, R. & Jones, J. D. Role of plant hormones in plant defence responses. Plant Mol. Biol. 69, 473–488 (2009).
Tsuda, K., Sato, M., Stoddard, T., Glazebrook, J. & Katagiri, F. Network properties of robust immunity in plants. PLoS Genet. 5, e1000772 (2009). A network approach to plant immunity shows complex interactions between defence hormone signalling pathways acting in both PTI and ETI.
Lipka, V. et al. Pre- and postinvasion defenses both contribute to nonhost resistance in Arabidopsis. Science 310, 1180–1183 (2005).
Stein, M. et al. Arabidopsis PEN3/PDR8, an ATP binding cassette transporter, contributes to nonhost resistance to inappropriate pathogens that enter by direct penetration. Plant Cell 18, 731–746 (2006).
Bednarek, P. et al. A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science 323, 101–106 (2009).
Clay, N. K., Adio, A. M., Denoux, C., Jander, G. & Ausubel, F. M. Glucosinolate metabolites required for an Arabidopsis innate immune response. Science 323, 95–101 (2009).
Krattinger, S. G. et al. A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science 323, 1360–1363 (2009). The first cloning of a broad spectrum resistance gene in wheat that is active against rusts and mildews. This study opened the way for the genetic manipulation of crop cultivars.
Wang, W., Wen, Y., Berkey, R. & Xiao, S. Specific targeting of the Arabidopsis resistance protein RPW8.2 to the interfacial membrane encasing the fungal Haustorium renders broad-spectrum resistance to powdery mildew. Plant Cell 21, 2898–2913 (2009).
Anelli, T. & Sitia, R. Protein quality control in the early secretory pathway. EMBO J. 27, 315–327 (2008).
Häweker, H. et al. Pattern recognition receptors require N-glycosylation to mediate plant immunity. J. Biol. Chem. 285, 4629–4636 (2010).
Li, J. et al. Specific ER quality control components required for biogenesis of the plant innate immune receptor EFR. Proc. Natl Acad. Sci. USA 106, 15973–15978 (2009).
Lu, X. et al. Uncoupling of sustained MAMP receptor signaling from early outputs in an Arabidopsis endoplasmic reticulum glucosidase II allele. Proc. Natl Acad. Sci. USA 106, 22522–22527 (2009).
Nekrasov, V. et al. Control of the pattern-recognition receptor EFR by an ER protein complex in plant immunity. EMBO J. 28, 3428–3438 (2009).
Saijo, Y. et al. Receptor quality control in the endoplasmic reticulum for plant innate immunity. EMBO J. 28, 3439–3449 (2009).
Vleeshouwers, V. G. A. A. et al. Effector genomics accelerates discovery and functional profiling of potato disease resistance and Phytophthora infestans avirulence genes. PLoS ONE 3, e2875 (2008).
Lacombe, S. et al. Interfamily transfer of a plant pattern-recognition receptor confers broad-spectrum bacterial resistance. Nature Biotech. 28, 365–369 (2010). A groundbreaking paper showing the potential for interfamily transfer of PRRs to provide broad spectrum disease protection in crop species.
Acknowledgements
We apologize to those authors whose workcouldnotbecited owing to space limitations. J.P.R. is an Australian Research Council Future Fellow. Work in P.N.D.'s laboratory is funded by the Australian Research Council, the US National Institutes of Health and the Grains Research and Development Corporation. We thank J. Ellis and B. Staskawicz for helpful discussions.
Author information
Authors and Affiliations
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Related links
DATABASES
TAIR
FURTHER INFORMATION
Glossary
- Elicitors
-
Molecules that induce ('elicit') an immune defence response. In the context of this Review, this term is used to refer to both pathogen-associated molecular patterns (PAMPs) and effectors.
- Pathogen-associated molecular patterns
-
Any of a number of conserved, usually structural, molecules common to pathogen organisms.
- Pattern recognition receptors
-
Plasma membrane-localized receptors that recognize the presence of pathogen-associated molecular patterns (PAMPs) in the extracellular environment.
- PAMP-triggered immunity
-
The plant defence response elicited by pathogen-associated molecular pattern (PAMP) recognition.
- Effectors
-
Proteins secreted by pathogens into host cells to enhance infection. Many of these function to suppress PAMP-triggered immunity responses.
- Effector-triggered immunity
-
The plant defence response elicited by effector recognition.
- Biotrophic
-
Biotrophic pathogens propagate in living plant tissue and generally do not cause necrosis as a result of infection. They use various means, such as haustoria production, to extract nutrients from host cells.
- Necrotrophic
-
Necrotrophic pathogens actively induce necrosis in infected tissues, often through the production of toxins, and obtain nutrients from the dead host tissue.
- Type-III secretion system
-
A syringe-like structure produced by many plant and animal pathogen bacteria that allows direct secretion of effector proteins from the bacterial cytoplasm into host cells.
- Haustoria
-
(sing. haustorium.) Specialized structures produced by some fungal and oomycete pathogens. Haustoria extend through the plant cell wall and expand in the host cell. They remain surrounded by a host-derived membrane and hence are topologically extracellular and separated from the host cytoplasm.
- Hemibiotrophic
-
Hemibiotrophic pathogens incorporate aspects of both biotrophic and necrotrophic infection strategies. Often this involves an initial biotrophic infection phase during which the pathogen spreads in host tissue, followed by a necrotrophic phase during which host cell death is induced.
- NB-LRR proteins
-
A class of intracellular receptor proteins containing nucleotide-binding (NB) and leucine-rich repeat (LRR) domains that recognize specific pathogen effectors.
Rights and permissions
About this article
Cite this article
Dodds, P., Rathjen, J. Plant immunity: towards an integrated view of plant–pathogen interactions. Nat Rev Genet 11, 539–548 (2010). https://doi.org/10.1038/nrg2812
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrg2812
This article is cited by
-
Magnaporthe oryzae effector AvrPik-D targets a transcription factor WG7 to suppress rice immunity
Rice (2024)
-
Host-pathogen interaction between pitaya and Neoscytalidium dimidiatum reveals the mechanisms of immune response associated with defense regulators and metabolic pathways
BMC Plant Biology (2024)
-
Commensal lifestyle regulated by a negative feedback loop between Arabidopsis ROS and the bacterial T2SS
Nature Communications (2024)
-
Targeting Magnaporthe oryzae effector MoErs1 and host papain-like protease OsRD21 interaction to combat rice blast
Nature Plants (2024)
-
Effects of plant tissue permeability on invasion and population bottlenecks of a phytopathogen
Nature Communications (2024)