Key Points
-
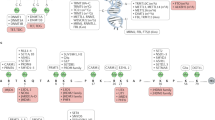
DNA methylation is an epigenetic modification of DNA that is important for the normal regulation of transcription, embryonic development, genomic imprinting, genome stability and chromatin structure.
-
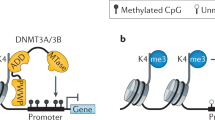
DNA methylation is controlled by DNA methyltransferases, methyl-CpG binding proteins and other chromatin-remodelling factors.
-
Aberrations in the DNA methylation system have an important role in human disease.
-
DNA methylation patterns are globally disrupted in cancer, with genome-wide hypomethylation and gene-specific hypermethylation events occurring simultaneously in the same cell.
-
Loss of normal imprinting contributes to several inherited genetic diseases in humans. These diseases include Beckwith–Wiedemann, Prader–Willi, and Angelman syndromes, Albright hereditary osteodystrophy (AHO) and pseudohypoparathyroidism Ia (PHP-Ia) and PHP-Ib, and transient neonatal diabetes.
-
In vitro manipulation of embryos during assisted reproduction procedures might lead to imprinting defects in the offspring.
-
Abnormal expansion of a CGG repeat in the FMR1 gene, accompanied by its hypermethylation and silencing, leads to fragile X syndrome. By contrast, contraction and hypomethylation of a larger 3.3 kb repeat leads to facioscapulohumeral muscular dystrophy.
-
Mutations in the machinery that regulates DNA methylation patterns and chromatin structure also contribute to human disease. Mutations in DNMT3B and ATRX lead to immunodeficiency, centromeric instability and facial anomalies (ICF) syndrome and Alpha-thalassemia/mental retardation syndrome, X-linked syndrome, respectively. Both disorders are characterized by localized disruptions in DNA methylation patterns.
-
The insulator/boundary proteins CCCTC-binding factor and BORIS, and the repression system known as RNAi, are probably involved in establishing and maintaining normal DNA methylation patterns.
Abstract
DNA methylation is a crucial epigenetic modification of the genome that is involved in regulating many cellular processes. These include embryonic development, transcription, chromatin structure, X chromosome inactivation, genomic imprinting and chromosome stability. Consistent with these important roles, a growing number of human diseases have been found to be associated with aberrant DNA methylation. The study of these diseases has provided new and fundamental insights into the roles that DNA methylation and other epigenetic modifications have in development and normal cellular homeostasis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Bird, A. DNA methylation patterns and epigenetic memory. Genes. Dev. 16, 6–21 (2002). An excellent review of DNA methylation in mammalian cells.
Fischle, W., Wang, Y. & Allis, C. D. Binary switches and modification cassettes in histone biology and beyond. Nature 425, 475–479 (2003).
Robertson, K. D. DNA methylation and chromatin — unraveling the tangled web. Oncogene 21, 5361–5379 (2002).
Hendrich, B. & Tweedie, S. The methyl-CpG binding domain and the evolving role of DNA methylation in animals. Trends Genet. 19, 269–277 (2003).
Mayer, W., Niveleau, A., Walter, J., Fundele, R. & Haaf, T. Demethylation of the zygotic paternal genome. Nature 403, 501–502 (2000). This paper demonstrates that active demethylation occurs during embryonic development in the absence of cell division.
Yoder, J. A., Walsh, C. P. & Bestor, T. H. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 13, 335–340 (1997).
Bird, A. CpG-rich islands and the function of DNA methylation. Nature 321, 209–213 (1986).
Song, F. et al. Association of tissue-specific differentially methylated regions (TDMs) with differential gene expression. Proc. Natl Acad. Sci. USA 102, 3336–3341 (2005).
Panning, B. & Jaenisch, R. DNA hypomethylation can activate Xist expression and silence X-linked genes. Genes. Dev. 10, 1991–2002 (1996).
Li, E., Bestor, T. H. & Jaenisch, R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69, 915–926 (1992).
Okano, M., Bell, D. W., Haber, D. A. & Li, E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99, 247–257 (1999). References 10 and 11 describe in detail the detrimental effects that loss of DNA methylation has on mouse embryonic development.
Brown, R. & Strathdee, G. Epigenomics and epigenetic therapy of cancer. Trends Mol. Med. 8 (Suppl.), S43–S48 (2002).
Novina, C. D. & Sharp, P. A. The RNAi revolution. Nature 430, 161–164 (2004).
Shahbazian, M. D. & Zoghbi, H. Y. Molecular genetics of Rett syndrome and clinical spectrum of MECP2 mutations. Curr. Opin. Neurol. 14, 171–176 (2001).
Feinberg, A. P. & Tycko, B. The history of cancer epigenetics. Nature Rev. Cancer 4, 1–11 (2004).
Mertens, F., Johansson, B., Hoglund, M. & Mitelman, F. Chromosomal imbalance maps of malignant solid tumors: A cytogenetic survey of 3185 neoplasms. Cancer Res. 57, 2765–2780 (1997).
Robertson, K. D. & Wolffe, A. P. DNA methylation in health and disease. Nature Rev. Genet. 1, 11–19 (2000).
Widschwendter, M. et al. DNA hypomethylation and ovarian cancer biology. Cancer Res. 64, 4472–4480 (2004).
De Smet, C., Lurquin, C., Lethe, B., Martelange, V. & Boon, T. DNA methylation is the primary silencing mechanism for a set of germ line- and tumor-specific genes with a CpG-rich promoter. Mol. Cell. Biol. 19, 7327–7335 (1999).
Nakamura, N. & Takenaga, K. Hypomethylation of the metastasis-associated S100A4 gene correlates with gene activation in human colon adenocarcinoma cell lines. Clin. Exp. Metastasis 16, 471–479 (1998).
Akiyama, Y., Maesawa, C., Ogasawara, S., Terashima, M. & Masuda, T. Cell-type specific repression of the maspin gene is disrupted frequently by demethylation at the promoter region in gastric intestinal metaplasia and cancer cells. Am. J. Clin. Pathol. 163, 1911–1919 (2003).
Gupta, A., Godwin, A. K., Vanderveer, L., Lu, A. & Liu, J. Hypomethylation of the synuclein γ gene CpG island promotes its aberrant expression in breast and ovarian carcinoma. Cancer Res. 63, 664–673 (2003).
Strichman-Almashanu, L. Z. et al. A genome-wide screen for normally methylated human CpG islands that can identify novel imprinted genes. Gen. Res. 12, 543–554 (2002).
Issa, J. -P. CpG island methylator phenotype in cancer. Nature Rev. Cancer 4, 988–993 (2004).
Chan, A. O.- O. et al. CpG island methylation in aberrant crypt foci of the colorectum. Am. J. Pathol. 160, 1823–1830 (2002).
Costello, J. F. et al. Aberrant CpG-island methylation has a non-random and tumor-type-specific patterns. Nature Genet. 25, 132–138 (2000). This paper represents one of the most comprehensive genome-wide scans for aberrant methylation in cancer and demonstrates just how widespread the defects are.
Laird, P. W. The power and the promise of DNA methylation markers. Nature Rev. Cancer 3, 253–266 (2003).
Feinberg, A. P., Cui, H. & Ohlsson, R. DNA methylation and genomic imprinting: insights from cancer into epigenetic mechanisms. Sem. Canc. Biol. 12, 389–398 (2002).
Reik, W. & Walter, J. Genomic imprinting: parental influence on the genome. Nature Rev. Genet. 2, 21–32 (2001).
Klenova, E. M., Morse, H. C., Ohlsson, R. & Lobanenkov, V. V. The novel BORIS + CTCF gene family is uniquely involved in the epigenetics of normal biology and cancer. Sem. Canc. Biol. 449, 1–16 (2002).
Kanduri, C. et al. Functional association of CTCF with the insulator upstream of the H19 gene is parent of origin-specific and methylation-sensitive. Curr. Biol. 10, 853–856 (2000). This paper shows that CTCF binding is methylation sensitive and provides important mechanistic insights into how cells distinguish between maternal and paternal alleles.
Thakur, N. et al. An antisense RNA regulates the bidirectional silencing property of the Kcnq1 imprinting control region. Mol. Cell. Biol. 24, 7855–7862 (2004).
Schoenherr, C. J., Levorse, J. M. & Tilghman, S. M. CTCF maintains differential methylation at the Igf2/H19 locus. Nature Genet. 33, 66–69 (2003).
Khosla, S., Dean, W., Brown, D., Reik, W. & Feil, R. Culture of preimplantation mouse embryos affects fetal development and the expression of imprinted genes. Biol. Reprod. 64, 918–926 (2001).
Behr, B. & Wang, H. Effects of culture conditions on IVF outcome. Eur. J. Obstet. Gynecol. Reprod. Biol. 115 (Suppl. 1), S72–S76 (2004).
Dean, W. et al. Conservation of methylation reprogramming in mammalian development: aberrant reprogramming in cloned embryos. Proc. Natl Acad. Sci. USA 98, 13734–13738 (2001).
De Baun, M. R., Niemitz, E. L. & Feinberg, A. P. Association of in vitro fertilization with Beckwith–Wiedemann syndrome and epigenetic alterations of LIT1 and H19. Am. J. Hum. Genet. 72, 156–160 (2003). References 34–37 show that in vitro culturing of embryos can lead to epigenetic defects in animals and that this might also have a role in humans.
Hao, Y., Crenshaw, T., Moulton, T., Newcomb, E. & Tycko, B. Tumour-suppressor activity of H19 RNA. Nature 365, 764–767 (1993).
Moulton, T. et al. Epigenetic lesions at the H19 locus in Wilms' tumor patients. Nature Genet. 7, 440–447 (1994).
Steenman, M. J. C. et al. Loss of imprinting of IGF2 is linked to reduced expression and abnormal methylation of H19 in Wilms' tumor. Nature Genet. 7, 433–439 (1994).
Sakatani, T. et al. Loss of imprinting of Igf2 alters intestinal maturation and tumorigenesis in mice. Science 307, 1976–1978 (2005).
Cui, H. et al. Loss of imprinting of insulin-like growth factor-II in Wilms' tumor commonly involves altered methylation but not mutations of CTCF or its binding site. Cancer Res. 61, 4947–4950 (2001).
Nakagawa, H. et al. Loss of imprinting of the insulin-like growth factor II gene occurs by biallelic methylation in a core region of H19-associated CTCF-binding sites in colorectal cancer. Proc. Natl Acad. Sci. USA 98, 591–596 (2001).
Cui, H. et al. Loss of imprinting in colorectal cancer linked to hypomethylation of H19 and IGF2. Cancer Res. 62, 6442–6446 (2002).
Thompson, J. S., Reese, K. J., DeBaun, M. R., Perlman, E. J. & Feinberg, A. P. Reduced expression of the cyclin-dependent kinase inhibitor gene p57KIP2 in Wilms' tumor. Cancer Res. 56, 5723–5727 (1996).
Yu, Y. et al. NOEY2 (ARHI), an imprinted putative tumor suppressor gene in ovarian and breast carcinomas. Proc. Natl Acad. Sci. USA 96, 214–219 (1999).
Pedersen, I. S. et al. Frequent loss of imprinting of PEG1/MEST in invasive breast cancer. Cancer Res. 59, 5449–5451 (1999).
Walter, J. & Paulsen, M. Imprinting and disease. Semin. Cell Dev. Biol. 14, 101–110 (2003).
Lee, M. P. et al. Loss of imprinting of a paternally expressed transcript, with antisense orientation to KvLQT1, occurs frequently in Beckwith–Wiedemann syndrome and is independent of insulin-like growth factor II imprinting. Proc. Natl Acad. Sci. USA 96, 5203–5208 (1999).
Kanduri, C. et al. A differentially methylated imprinting control region within the Kcnq1 locus harbors a methylation-sensitive chromatin insulator. J. Biol. Chem. 277, 18106–18110 (2002).
Butler, M. G. Imprinting disorders: non-Mendelian mechanisms affecting growth. J. Pediatr. Endocrinol. Metab. 15, 1279–1288 (2002).
Weksberg, R. et al. Tumor development in the Beckwith–Wiedemann syndrome is associated with a variety of constitutional molecular 11p15 alterations including imprinting defects of KCNQ1OT1. Hum. Mol. Genet. 10, 2989–3000 (2001).
Diaz-Meyer, N. et al. Silencing of CDKN1C (p57KIP2) is associated with hypomethylation at KvDMR1 in Beckwith–Wiedemann syndrome. J. Med. Genet. 40, 797–801 (2005).
Goldstone, A. P. Prader–Willi syndrome: advances in genetics, pathophysiology and treatment. Trends Endocrinol. Metab. 15, 12–20 (2004).
Nicholls, R. D. & Knepper, J. L. Genome organization, function, and imprinting in Prader–Willi and Angelman syndromes. Annu. Rev. Genomics Hum. Genet. 2, 153–175 (2001).
Sutcliffe, J. S. et al. Deletions of a differentially methylated CpG island at the SNRPN gene define a putative imprinting control region. Nature Genet. 8, 52–58 (1994).
Runte, M. et al. Comprehensive methylation analysis in typical and atypical PWS and AS patients with normal biparental chromosomes 15. Eur. J. Hum. Genet. 9, 519–526 (2001).
Runte, M. et al. The IC-SNURF–SNRPN transcript serves as a host for multiple small nucleolar RNA species and as an antisense RNA for UBE3A. Hum. Mol. Genet. 10, 2687–2700 (2001).
Gerard, G., Hernandez, L., Wevrick, R. & Stewart, C. L. Disruption of the mouse necdin gene results in early post-natal lethality. Nature Genet. 23, 199–202 (1999).
Tsai, T. F., Jiang, Y. H., Bressler, J., Armstrong, D. & Beaudet, A. L. Paternal deletion from Snrpn to Ube3a in the mouse causes hypotonia, growth retardation and partial lethality and provides evidence for a gene contributing to Prader–Willi syndrome. Nature Genet. 8, 1357–1364 (1999).
Saitoh, S. & Wada, T. Parent-of-origin specific histone acetylation and reactivation of a key imprinted gene locus in Prader–Willi syndrome. Am. J. Hum. Genet. 66, 1958–1962 (2000).
Lossie, A. C. et al. Distinct phenotypes distinguish the molecular classes of Angelman syndrome. J. Med. Genet. 38, 834–845 (2001).
Buiting, K., Lich, C., Cottrell, S., Barnicoat, A. & Horsthemke, B. A 5-kb imprinting center deletion in a family with Angelman syndrome reduces the shortest region of deletion overlap to 880 bp. Hum. Genet. 105, 665–666 (1999).
Lalande, M. Imprints of disease at GNAS1. J. Clin. Inves. 107, 793–794 (2001).
Liu, J., Nealon, J. G. & Weinstein, L. S. Distinct patterns of abnormal GNAS imprinting in familial and sporadic pseudohypoparathyroidism type IB. Hum. Mol. Genet. 14, 95–102 (2005).
Hayward, B. E. et al. Imprinting of the G s α gene GNAS1 in the pathology of acromegaly. J. Clin. Inves. 107, R31–R36 (2001).
Temple, I. K. & Shield, J. P. H. Transient neonatal diabetes, a disorder of imprinting. J. Med. Genet. 39, 872–875 (2002).
Arima, T. et al. A conserved imprinting control region at the HYMAI/ZAC domain is implicated in transient neonatal diabetes mellitus. Hum. Mol. Genet. 10, 1475–1483 (2001).
Morison, I. M. & Reeve, A. E. A catalogue of imprinted genes and parent-of-origin effects in humans and animals. Hum. Mol. Genet. 7, 1599–1609 (1998). An excellent review detailing all known disorders in humans and animals that have a parent-of-origin bias.
Lewis, A. & Murrell, A. Genomic imprinting: CTCF protects the boundaries. Curr. Biol. 14, R284–R286 (2004).
Everett, C. M. & Wood, N. W. Trinucleotide repeats and neurodegenerative disease. Brain 127, 2385–2405 (2004).
Cleary, J. D. & Pearson, C. E. The contribution of cis-elements to disease-associated repeat instability: clinical and experimental evidence. Cytogenet. Genome Res. 100, 25–55 (2003).
Crawford, D. C., Acuna, J. M. & Sherman, S. L. FMR1 and the fragile X syndrome: human genome epidemiology review. Genet. Med. 3, 359–371 (2001).
Sutherland, G. R. Rare fragile sites. Cytogenet. Genome Res. 100, 77–84 (2003).
Jin, P., Alisch, R. S. & Warren, S. T. RNA and microRNAs in fragile X mental retardation. Nature Cell Biol. 6, 1048–1053 (2004).
Hagerman, R. J. et al. Intention tremor, parkinsonism, and generalized brain atrophy in male carriers of fragile X. Neurology 57, 127–130 (2001).
Oberle, I. et al. Instability of a 550-base pair segment and abnormal methylation in fragile X syndrome. Science 252, 1097–1102 (1991).
Coffee, B., Zhang, F., Ceman, S., Warren, S. T. & Reines, D. Histone modifications depict an aberrantly heterochromatinized FMR1 gene in fragile X syndrome. Am. J. Hum. Genet. 71, 923–932 (2002).
Sarafidou, T. et al. Folate-sensitive fragile site FRA10A is due to an expansion of a CGG repeat in a novel gene, FRA10AC1, encoding a nuclear protein. Genomics 84, 69–81 (2004).
van Overveld, P. G. M. et al. Hypomethylation of D4Z4 in 4q-linked and non-4q-linked facioscapulohumeral muscular dystrophy. Nature Genet. 34, 315–317 (2003).
Rijkers, T. et al. FRG2, an FSHD candidate gene, is transcriptionally upregulated in differentiating primary myoblast cultures of FSHD patients. J. Med. Genet. 41, 826–836 (2004).
Gabellini, D., Green, M. R. & Tupler, R. Inappropriate gene activation in FSHD: a repressor complex binds a chromosomal repeat deleted in dystrophic muscle. Cell 110, 339–348 (2002).
Jiang, G. et al. Testing the position-effect variegation hypothesis for facioscapulohumeral muscular dystrophy by analysis of histone modification and gene expression in subtelomeric 4q. Hum. Mol. Genet. 12, 2909–2921 (2003).
Steinbach, P., Glaser, D., Vogel, W., Wolf, M. & Schwemmle, S. The DMPK gene of severely affected myotonic dystrophy patients is hypermethylated proximal to the largely expanded CTG repeat. Am. J. Hum. Genet. 62, 278–285 (1998).
Filippova, G. N. et al. CTCF-binding sites flank CTG/CAG repeats and form a methylation-sensitive insulator at the DM1 locus. Nature Genet. 28, 335–343 (2001).
Saveliev, A., Everett, C., Sharpe, T., Webster, Z. & Festenstein, R. DNA triplet repeats mediate heterochromatin-protein-1-sensitive variegated gene silencing. Nature 422, 909–913 (2003).
Edamura, K. N., Leonard, M. R. & Pearson, C. E. Role of replication and CpG methylation in fragile X syndrome CGG deletions in primate cells. Am. J. Hum. Genet. 76, 302–311 (2005).
Gorbunova, V., Seluanov, A., Mittelman, D. & Wilson, J. H. Genome-wide demethylation destabilizes CTG•CAG trinucleotide repeats in mammalian cells. Hum. Mol. Genet. 13, 2979–2989 (2004).
Reik, W., Maher, E. R., Morrison, P. J., Narding, A. E. & Simpson, S. A. Age of onset in Huntington's disease and methylation at D4S95. J. Med. Genet. 30, 185–188 (1993).
Smith, S. S., Laayoun, A., Lingeman, R. G., Baker, D. J. & Riley, J. Hypermethylation of telomere-like foldbacks at codon 12 of the human c-Ha-ras gene and the trinucleotide repeat of the FMR-1 gene of fragile X. J. Mol. Biol. 243, 143–151 (1994).
Handa, V., Saha, T. & Usdin, K. The fragile X syndrome repeats form RNA hairpins that do not activate the interferon-inducible protein kinase, PKR, but are cut by dicer. Nucleic Acids Res. 31, 6243–6248 (2003).
Klippel, J. H. Systemic lupus erythematosus: demographics, prognosis, and outcome. J. Rheumatol. 24, 67–71 (1997).
Richardson, B. DNA methylation and autoimmune disease. Clin. Immunol. 109, 72–79 (2003).
Richardson, B. et al. Evidence for impaired T cell DNA methylation in systemic lupus erythematosus and rheumatoid arthritis. Arthritis Rheum. 33, 1665–1673 (1990).
Quddus, J. et al. Treating activated CD4+ T cells with either of two distinct DNA methyltransferase inhibitors, 5-azacytidine or procainamide, is sufficient to cause a lupus-like disease in syngeneic mice. J. Clin. Inves. 92, 38–53 (1993).
Lu, Q. et al. Demethylation of ITGAL (CD11a) regulatory sequences in systemic lupus erythematosus. Arthritis Rheum. 46, 1282–1291 (2003).
Oelke, K. et al. Overexpression of CD70 and overstimulation of IgG synthesis by lupus T cells and T cells treated with DNA methylation inhibitors. Arthritis Rheum. 50, 1850–1860 (2004).
Sekigawa, I. et al. DNA methylation in systemic erythematosus. Lupus 12, 79–85 (2003).
Scheinbart, L. S., Johnson, M. A., Gross, L. A., Edelstein, S. R. & Richardson, B. C. Procainamide inhibits DNA methyltransferase in a human T cell line. J. Rheum. 18, 530–540 (1991).
Deng, C. et al. Hydralazine may induce autoimmunity by inhibiting extracellular signal-regulated kinase pathway signalling. Arthritis Rheum. 48, 746–756 (2003).
Smeets, D. F. et al. ICF syndrome: a new case and review of the literature. Hum. Genet. 94, 240–246 (1994).
Ehrlich, M. The ICF syndrome, a DNA methyltransferase 3B deficiency and immunodeficiency disease. Clin. Immunol. 109, 17–28 (2003).
Franceschini, P. et al. Variability of clinical and immunological phenotype in immunodeficiency-centromeric instability-facial anomalies syndrome. Eur. J. Pediatr. 154, 840–846 (1995).
Tuck-Muller, C. M. et al. DNA hypomethylation and unusual chromosome instability in cell lines from ICF patients. Cytogenet. Cell Genet. 89, 121–128 (2000).
Xu, G. -L. et al. Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature 402, 187–191 (1999).
Hansen, R. S. et al. The DNMT3B DNA methyltransferase gene is mutated in the ICF immunodeficiency syndrome. Proc. Natl. Acad. Sci. USA 96, 14412–14417 (1999).
Gowher, H. & Jeltsch, A. Molecular enzymology of the catalytic domains of the Dnmt3a and Dnmt3b DNA methyltransferases. J. Biol. Chem. 277, 20409–20414 (2002).
Jiang, Y. L. et al. DNMT3B mutations and DNA methylation defect define two types of ICF syndrome. Hum. Mut. 25, 56–63 (2005).
Kondo, T. et al. Whole-genome methylation scan in ICF syndrome: hypomethylation of non-satellite DNA repeats D4Z4 and NBL2. Hum. Mol. Genet. 9, 597–604 (2000).
Tao, Q. et al. Defective de novo methylation of viral and cellular DNA sequences in ICF syndrome cells. Hum. Mol. Genet. 11, 2091–2102 (2002).
Hansen, R. S. et al. Escape from gene silencing in ICF syndrome: evidence for advanced replication time as a major determinant. Hum. Mol. Genet. 9, 2575–2587 (2000).
Ehrlich, M. DNA hypomethylation, cancer, the immunodeficiency, centromeric region instability, facial anomalies syndrome and chromosomal rearrangements. J. Nutr. 132, S2424–S2429 (2002).
Ehrlich, M. et al. DNA methyltransferase 3B mutations linked to the ICF syndrome cause dysregulation of lymphomagenesis genes. Hum. Mol. Genet. 10, 2917–2931 (2001).
Geiman, T. M. et al. DNMT3B interacts with hSNF2H chromatin remodeling enzyme, HDACs 1 and 2, and components of the histone methylation system. Biochem. Biophys. Res. Commun. 318, 544–555 (2004).
Geiman, T. M. et al. Isolation and characterization of a novel DNA methyltransferase complex linking DNMT3B with components of the mitotic chromosome condensation machinery. Nucleic Acids Res. 32, 2716–2729 (2004).
Bai, S. et al. DNA methyltransferase 3b regulates nerve growth factor-induced differentiation of PC12 cells by recruiting histone deacetylase 2. Mol. Cell. Biol. 25, 751–766 (2005).
Havas, K., Whitehouse, I. & Owen-Hughes, T. ATP-dependent chromatin remodeling activities. Cell. Mol. Life Sci. 58, 673–682 (2001).
Gibbons, R. J. et al. Mutations in transcriptional regulator ATRX establish the functional significance of a PHD-like domain. Nature Genet. 17, 146–148 (1997).
Cardoso, C. et al. ATR-X mutations cause impaired nuclear location and altered DNA binding properties of the XNP/ATR-X protein. J. Med. Genet. 37, 746–751 (2000).
Tang, J. et al. A novel transcription regulatory complex containing death domain-associated protein and the ATR-X syndrome protein. J. Biol. Chem. 279, 20369–20377 (2004).
Ishov, A. M., Vlafimirova, O. V. & Maul, G. G. Heterochromatin and ND10 are cell-cycle regulated and phosphorylation-dependent alternate nuclear sites of the transcription repressor Daxx and SWI/SNF protein ATRX. J. Cell. Sci. 117, 3807–3820 (2004).
Xue, Y. et al. The ATRX syndrome protein forms a chromatin-remodeling complex with Daxx and localizes in promyelocytic leukemia nuclear bodies. Proc. Natl Acad. Sci. USA 100, 10635–10640 (2003).
Gibbons, R. J. et al. Mutations in ATRX, encoding a SWI/SNF-like protein, cause diverse changes in the patterns of DNA methylation. Nature Genet. 24, 368–371 (2000).
Jones, P. A. & Baylin, S. B. The fundamental role of epigenetic events in cancer. Nature Rev. Genet. 3, 415–428 (2002).
Filippova, G. N. et al. Boundaries between chromosomal domains of X inactivation and escape bind CTCF and lack CpG methylation during early development. Dev. Cell 8, 31–42 (2005).
Loukinov, D. I. et al. BORIS, a novel male germ-line-specific protein associated with epigenetic reprogramming events, shares the same 11-zinc-finger domain with CTCF, the insulator protein involved in reading imprinting marks in the soma. Proc. Natl Acad. Sci. USA 99, 6806–6811 (2002). The first in-depth characterization of the CTCF paralogue BORIS and a demonstration of the mutually exclusive expression of these two proteins in normal tissues.
Verdel, A. et al. RNAi-mediated targeting of heterochromatin by the RITS complex. Science 303, 672–676 (2004).
Noma, K. -i. et al. RITS acts in cis to promote RNA interference-mediated transcriptional and post-transcriptional silencing. Nature Genet. 36, 1174–1180 (2004).
Schramke, V. & Allshire, R. Hairpin RNAs and retrotransposon LTRs effect RNAi and chromatin-based gene silencing. Science 301, 1069–1073 (2003).
Motamedi, M. R. et al. Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell 119, 789–802 (2004). References 127–130 provided the first direct, mechanistic evidence for how the RNAi machinery can target transcriptional repression back to heterochromatic repeat regions of the genome.
Kawasaki, H. & Taira, K. Induction of DNA methylation and gene silencing by short interfering RNAs in human cells. Nature 431, 211–217 (2004).
Rudert, F., Bronner, S., Garnier, J. M. & Dolle, P. Transcripts from opposite strands of γ satellite DNA are differentially expressed during mouse development. Mamm. Genome 6, 76–83 (1995).
Lehnertz, B. et al. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr. Biol. 13, 1192–1200 (2003).
Tufarelli, C. et al. Transcription of antisense RNA leading to gene silencing and methylation as a novel cause of human genetic disease. Nature Genet. 34, 157–165 (2003).
Shendure, J. & Church, G. M. Computational discovery of sense–antisense transcription in the human and mouse genomes. Genome Biol. 3, 1–14 (2003).
Lavorgna, G. et al. In seach of antisense. Trends Biochem. Sci. 29, 88–94 (2004).
Mukhopadhyay, R. et al. The binding sites for the chromatin insulator protein CTCF map to DNA methylation-free domains genome-wide. Gen. Res. 14, 1594–1602 (2004).
Fedoriw, A. M., Stein, P., Svoboda, P., Schultz, R. M. & Bartolomei, M. S. Transgenic RNAi reveals essential function for CTCF in H19 gene imprinting. Science 303, 238–240 (2004).
Rand, E., Ben-Porath, I., Keshet, I. & Cedar, H. CTCF elements direct allele-specific undermethylation at the imprinted H19 locus. Curr. Biol. 14, 1007–1012 (2004).
Rasko, J. E. J. et al. Cell growth inhibition by the multifunctional multivalent zinc-finger factor CTCF. Cancer Res. 61, 6002–6007 (2001).
Filippova, G. N. et al. Tumor-associated zinc finger mutations in the CTCF transcription factor selectively alter its DNA-binding specificity. Cancer Res. 62, 48–52 (2002).
Wright, V. C., Schieve, L. A., Reynolds, M. A., Jeng, G. & Kissin, D. Assisted reproductive technology surveillance — United States, 2001. MMWR Surveill Summ. 53, 1–20 (2004).
Cox, G. F. et al. Intracytoplasmic sperm injection may increase the risk of imprinting defects. Am. J. Hum. Genet. 71, 162–164 (2002).
Gicquel, C. et al. In vitro fertilization may increase the risk of Beckwith–Wiedemann syndrome related to the abnormal imprinting of the KCN1OT gene. Am. J. Hum. Genet. 72, 1338–1341 (2003).
Reik, W., Dean, W. & Walter, J. Epigenetic reprogramming in mammalian development. Science 293, 1089–1093 (2001).
Li, E., Beard, C. & Jaenisch, R. Role for DNA methylation in genomic imprinting. Nature 366, 362–365 (1993).
De Chiara, T. M., Robertson, E. J. & Efstratiadis, A. Parental imprinting of the mouse insulin-like growth factor II gene. Cell 64, 849–859 (1991).
Barker, D. J. et al. Fetal nutrition and cardiovascular disease in adult life. Lancet 341, 938–941 (1993).
Mann, M. R. W. et al. Selective loss of imprinting in the placenta following preimplantation development in culture. Development 131, 3727–3735 (2004).
Acknowledgements
I apologize to those whose work could not be cited owing to space limitations. Work in the author's lab is supported by the National Institutes of Health.
Author information
Authors and Affiliations
Ethics declarations
Competing interests
The author declares no competing financial interests.
Supplementary information
Supplementary information S1
(PDF 106 kb)
Supplementary information S2
(PDF 139 kb)
Supplementary information S3
(PDF 188 kb)
Related links
Related links
DATABASES
Entrez gene
Omim
Albright hereditary osteodystrophy
alpha-thalassemia/mental retardation syndrome, X-linked
facioscapulohumeral muscular dystrophy
immunodeficiency, centromeric instability and facial anomalies syndrome
transient neonatal diabetes mellitus
FURTHER INFORMATION
The M. D. Anderson Cancer Center DNA Methylation in Cancer web site
The Genomic Imprinting Website
MRC Mammalian Genetics Unit Mouse Imprinting web site
CITE: Candidate Imprinted Transcript from gene Expression database
Glossary
- CPG ISLAND
-
A genomic region of ∼1 kb that has a high G–C content, is rich in CpG dinucleotides and is usually hypomethylated.
- TRANSCRIPTIONAL INTERFERENCE
-
Repression of one transcriptional unit by another such unit that is linked in cis.
- RESTRICTION LANDMARK GENOMIC SCANNING
-
(RLGS). A genome-wide method for analyzing the DNA methylation status of CpG islands. Radiolabelled fragments obtained by digestion with NotI (a methylation-sensitive restriction enzyme) are separated by two-dimensional gel electrophoresis, allowing differentiation between methylated and unmethylated regions.
- ENHANCER
-
A regulatory DNA element that usually binds several transcription factors and can activate transcription from a promoter at great distance and in an orientation-independent manner.
- UNIPARENTAL DISOMY
-
Inheritance of a chromosome or chromosome region from a single parent.
- BALANCED TRANSLOCATION
-
A condition in which two pieces of chromosomal material have switched places, but the correct number of chromosomes has been maintained.
- UBIQUITIN–PROTEASOME DEGRADATION PATHWAY
-
Degradation pathway in which a protein that has been post-translationally modified with several ubiquitin polypeptides is targeted for destruction to the proteasome, a large cytosolic protein complex with several proteolytic activities.
- CHROMOGRANINS
-
A group of acidic, soluble, secretory proteins that are produced by neurons and neuroendocrine cells.
- OKAZAKI FRAGMENTS
-
Short pieces of DNA that are synthesized on the lagging strand at the replication fork.
- FOLATE-SENSITIVE FRAGILE SITE
-
A region of chromatin that fails to compact normally during mitosis and that can be observed after culturing cells in media that is deficient in folic acid and thymidine.
- RNAI
-
The process whereby double-stranded RNAs are cleaved into 21–23 nucleotide duplexes termed small interfering RNAs, leading to inhibition of expression of genes that contain a complementary sequence.
- DICER
-
A ribonuclease that processes dsRNAs to ∼21 nucleotide siRNAs (for RNAi) or excises microRNAs from their hairpin precursors.
- CD4+ T CELL
-
Also known as a helper T cell. Initiates both antibody production by B cells and stimulates the activation of other immune cells, such as macrophages, after recognizing a portion of a protein antigen on the surface of an antigen presenting cell.
- ADOPTIVE TRANSFER
-
The process of conferring immunity to an individual by transferring cells or serum from another individual that has been immunized with a specific antigen.
- SYNGENEIC MACROPHAGES
-
macrophages (immune cells that engulf foreign particles) that are transferred between genetically identical mice.
- LYMPHOBLASTOID CELL LINE
-
Immortalized B cell line created by infecting primary B cells with Epstein-Barr virus.
- SNF2-LIKE PROTEINS
-
ATP-dependent chromatin remodelling enzymes that contain a region homologous to an extended family of proteins that include known RNA and DNA helicases.
- PML BODIES
-
Dot-like structures in the nucleus of most mammalian cells. These were originally defined by the localization of the PML protein, which is involved in transcriptional regulation.
Rights and permissions
About this article
Cite this article
Robertson, K. DNA methylation and human disease. Nat Rev Genet 6, 597–610 (2005). https://doi.org/10.1038/nrg1655
Issue Date:
DOI: https://doi.org/10.1038/nrg1655
This article is cited by
-
5-Methylcytosine immunohistochemistry for predicting cutaneous melanoma prognosis
Scientific Reports (2024)
-
Obesity is associated with quality of sperm parameters in men with infertility: a cross-sectional study
Reproductive Health (2023)
-
DNMT1 SNPs (rs2114724 and rs2228611) associated with positive symptoms in Chinese patients with schizophrenia
Annals of General Psychiatry (2023)
-
Sex differences in the intergenerational link between maternal and neonatal whole blood DNA methylation: a genome-wide analysis in 2 birth cohorts
Clinical Epigenetics (2023)
-
Age-related promoter-switch regulates Runx1 expression in adult rat hearts
BMC Cardiovascular Disorders (2023)