Key Points
-
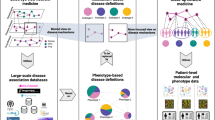
Disease progression patterns of patients with more than one disease have recently received increasing attention, as disease co-occurrences can help to elucidate the interaction between the molecular level and external exposures such as diet, lifestyle and patient care. Additionally, they can provide information about the underlying network biology of shared and multifunctional genes and pathways.
-
The concepts of pleiotropy, robustness and rewiring are central to the investigation of comorbidity and network dynamics and should be viewed together, as they all relate to the disease trajectory of an individual.
-
The temporal disease progression of the non-idealized patient can be described in terms of trajectories in a multimorbidity space, in which each dimension corresponds to a quantitative phenotype.
-
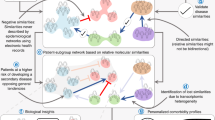
Dynamic network models can be constructed to study complex disease progression and are increasingly becoming feasible with the advances in high-throughput omics, single-cell technologies and sophisticated analysis tools.
-
The utility of network concepts has been hampered by confusion and inconsistent terminology. This can be mediated by the clear delineation of the concepts, especially in regards to context, including the clear specification of timeframe, phenotype and organizational level.
-
The increased collection of health transaction data combined with advances in omics technologies require a further concerted view on how robustness, rewiring and pleiotropy come together in frameworks that can rationalize comorbidities and their relationships at the molecular level, knowledge that can also facilitate drug repositioning and the development of targeted therapeutic strategies.
Abstract
The co-occurrence of diseases can inform the underlying network biology of shared and multifunctional genes and pathways. In addition, comorbidities help to elucidate the effects of external exposures, such as diet, lifestyle and patient care. With worldwide health transaction data now often being collected electronically, disease co-occurrences are starting to be quantitatively characterized. Linking network dynamics to the real-life, non-ideal patient in whom diseases co-occur and interact provides a valuable basis for generating hypotheses on molecular disease mechanisms, and provides knowledge that can facilitate drug repurposing and the development of targeted therapeutic strategies.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Tyson, J. J., Chen, K. & Novak, B. Network dynamics and cell physiology. Nat. Rev. Mol. Cell. Biol. 2, 908–916 (2001).
Paaby, A. B. & Rockman, M. V. The many faces of pleiotropy. Trends Genet. 29, 66–73 (2013).
Solovieff, N., Cotsapas, C., Lee, P. H., Purcell, S. M. & Smoller, J. W. Pleiotropy in complex traits: challenges and strategies. Nat. Rev. Genet. 14, 483–495 (2013). A review presenting the concept of pleiotropy and its controversies in the light of GWAS for complex traits.
Hodgkin, J. Seven types of pleiotropy. Int. J. Dev. Biol. 42, 501–505 (1998).
Pyeritz, R. E. Pleiotropy revisited: molecular explanations of a classic concept. Am. J. Med. Genet. 34, 124–134 (1989).
Wagner, G. P. & Zhang, J. The pleiotropic structure of the genotype–phenotype map: the evolvability of complex organisms. Nat. Rev. Genet. 12, 204–213 (2011).
Félix, M.-A. & Barkoulas, M. Pervasive robustness in biological systems. Nat. Rev. Genet. 16, 483–496 (2015). A comprehensive review of the concept of robustness.
Kitano, H. Towards a theory of biological robustness. Mol. Syst. Biol. 3, 137 (2007).
Diss, G. et al. Integrative avenues for exploring the dynamics and evolution of protein interaction networks. Curr. Opin. Biotechnol. 24, 775–783 (2013).
Ideker, T. & Krogan, N. J. Differential network biology. Mol. Syst. Biol. 8, 565 (2012). A review presenting the main approaches in differential network biology.
Lambert, J.-P. et al. Mapping differential interactomes by affinity purification coupled with data-independent mass spectrometry acquisition. Nat. Methods 10, 1239–1245 (2013).
Greene, C. S. et al. Understanding multicellular function and disease with human tissue-specific networks. Nat. Genet. 47, 569–576 (2015).
Bindea, G. et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity 39, 782–795 (2013).
Sun, S., Liu, Z., Zeng, T., Wang, Y. & Chen, L. Spatio-temporal analysis of type 2 diabetes mellitus based on differential expression networks. Sci. Rep. 3, 2268 (2013).
Bandyopadhyay, S. et al. Rewiring of genetic networks in response to DNA damage. Science 330, 1385–1389 (2010). The first study that uses differential epistasis mapping to systematically map out massive rewiring of genetic interaction networks in yeast.
Bensimon, A., Heck, A. J. & Aebersold, R. Mass spectrometry-based proteomics and network biology. Annu. Rev. Biochem. 81, 379–405 (2012).
Bisson, N. et al. Selected reaction monitoring mass spectrometry reveals the dynamics of signaling through the GRB2 adaptor. Nat. Biotechnol. 29, 653–658 (2011).
Lee, M. J. et al. Sequential application of anticancer drugs enhances cell death by rewiring apoptotic signaling networks. Cell 149, 780–794 (2012). This study monitors the rewiring of breast cancer cells to time- and order-dependent combinations of therapeutic agents and detects the optimal combination that can push cancer cells into a more vulnerable state.
Creixell, P. et al. Kinome-wide decoding of network attacking mutations driving cancer signaling. Cell 163, 202–217 (2015).
Gijsen, R. et al. Causes and consequences of comorbidity: a review. J. Clin. Epidemiol. 54, 661–674 (2001).
Von Lueder, T. G. & Atar, D. Comorbidities and polypharmacy. Heart Fail. Clin. 10, 367–372 (2014).
Feinstein, A. R. The pre-therapeutic classification co-morbidity in chronic disease. J. Chronic Dis. 23, 455–468 (1970).
Valderas, J. M., Sibbald, B. & Salisbury, C. Defining comorbidity: implications for understanding health and health services. Ann. Fam. Med. 7, 357–363 (2009).
Meghani, S. H. et al. The conceptualization and measurement of comorbidity: a review of the interprofessional discourse. Nurs. Res. Pract. 2013, 192782 (2013).
Loscalzo, J., Kohane, I. & Barabási, A.-L. Human disease classification in the postgenomic era: a complex systems approach to human pathobiology. Mol. Syst. Biol. 3, 124 (2007).
Maj, M. 'Psychiatric comorbidity': an artefact of current diagnostic systems? Br. J. Psychiatry 186, 182–184 (2005).
Radner, H., Yoshida, K., Smolen, J. S. & Solomon, D. H. Multimorbidity and rheumatic conditions-enhancing the concept of comorbidity. Nat. Rev. Rheumatol. 10, 252–256 (2014).
van den Akker, M., Buntinx, F. & Knottnerus, J. A. Comorbidity or multimorbidity. Eur. J. Gen. Pract. 2, 65–70 (2009).
Scanlon, P. H. Diabetic retinopathy. Medicine 38, 656–660 (2010).
Thomas, M. C. et al. Diabetic kidney disease. Nat. Rev. Dis. Prim. 1, 15038 (2015).
Barnes, P. J. et al. Chronic obstructive pulmonary disease. Nat. Rev. Dis. Prim. 362, 15076 (2015).
Mannino, D. & Kiri, V. Changing the burden of COPD mortality. Int. J. Chron. Obstruct. Pulmon. Dis. 1, 219–233 (2006).
Agustí, A. & Vestbo, J. Current controversies and future perspectives in chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 184, 507–513 (2012).
Hidalgo, C. A, Blumm, N., Barabási, A.-L. & Christakis, N. A. A dynamic network approach for the study of human phenotypes. PLoS Comput. Biol. 5, e1000353 (2009).
Roque, F. S. et al. Using electronic patient records to discover disease correlations and stratify patient cohorts. PLoS Comput. Biol. 7, e1002141 (2011).
Jensen, A. B. et al. Temporal disease trajectories condensed from population-wide registry data covering 6.2 million patients. Nat. Commun. 5, 4022 (2014). This paper uses registry data on 6.2 million patients from the Danish population to create temporal disease trajectories.
Oh, W. et al. Type 2 diabetes mellitus trajectories and associated risks. Big Data 4, 25–30 (2016).
Capobianco, E. & Liò, P. Comorbidity: a multidimensional approach. Trends Mol. Med. 19, 515–521 (2013).
Capobianco, E. & Liò, P. Comorbidity networks: beyond disease correlations. J. Complex. Networks 3, 319–332 (2015).
Gibson, G. Decanalization and the origin of complex disease. Nat. Rev. Genet. 10, 134–140 (2009).
Faner, R. et al. Molecular and clinical diseasome of comorbidities in exacerbated COPD patients. Eur. Respir. J. 46, 1001–1010 (2015).
Ibáñez, K., Boullosa, C., Tabarés-Seisdedos, R., Baudot, A. & Valencia, A. Molecular evidence for the inverse comorbidity between central nervous system disorders and cancers detected by transcriptomic meta-analyses. PLoS Genet. 10, e1004173 (2014).
Ellinghaus, D. et al. Analysis of five chronic inflammatory diseases identifies 27 new associations and highlights disease-specific patterns at shared loci. Nat. Genet. 48, 510–518 (2016). GWAS on five chronic inflammatory diseases detecting shared disease variants that could not have been found using a single disease approach.
Park, J., Lee, D.-S., Christakis, N. a & Barabási, A.-L. The impact of cellular networks on disease comorbidity. Mol. Syst. Biol. 5, 262 (2009). This paper combines Medicare clinical data and cellular OMIM data to assess the impact of cellular networks on comorbidity.
Catalá-López, F. et al. Inverse and direct cancer comorbidity in people with central nervous system disorders: a meta-analysis of cancer incidence in 577,013 participants of 50 observational studies. Psychother. Psychosom. 83, 89–105 (2014).
Driver, J. a et al. Inverse association between cancer and Alzheimer's disease: results from the Framingham Heart Study. BMJ 344, e1442 (2012).
Tabarés-Seisdedos, R. et al. No paradox, no progress: inverse cancer comorbidity in people with other complex diseases. Lancet. Oncol. 12, 604–608 (2011).
Pernicova, I. & Korbonits, M. Metformin—mode of action and clinical implications for diabetes and cancer. Nat. Rev. Endocrinol. 10, 143–156 (2014).
Kohane, I. S. Deeper, longer phenotyping to accelerate the discovery of the genetic architectures of diseases. Genome Biol. 15, 115 (2014).
Robinson, P. N. Deep phenotyping for precision medicine. Hum. Mutat. 33, 777–780 (2012).
Miotto, R., Li, L., Kidd, B. A. & Dudley, J. T. Deep patient: an unsupervised representation to predict the future of patients from the electronic health records. Sci. Rep. 6, 26094 (2016).
Zheng, X. et al. Genome-wide copy-number variation study of psychosis in Alzheimer's disease. Transl. Psychiatry 5, e574 (2015).
Plate, L. in Festschrift zum sechzigsten Geburtstag Richard Hertwigs. 536–610 (in German) (Fischer, 1910).
Sivakumaran, S. et al. Abundant pleiotropy in human complex diseases and traits. Am. J. Hum. Genet. 89, 607–618 (2011).
Rzhetsky, A., Wajngurt, D., Park, N. & Zheng, T. Probing genetic overlap among complex human phenotypes. Proc. Natl Acad. Sci. USA 104, 11694–11699 (2007).
Cotsapas, C. et al. Pervasive sharing of genetic effects in autoimmune disease. PLoS Genet. 7, e1002254 (2011).
Parkes, M., Cortes, A., van Heel, D. A. & Brown, M. A. Genetic insights into common pathways and complex relationships among immune-mediated diseases. Nat. Rev. Genet. 14, 661–673 (2013).
Ellis, J. D. et al. Tissue-specific alternative splicing remodels protein-protein interaction networks. Mol. Cell 46, 884–892 (2012). This paper mines 110 million electronic medical records and detects thousands of associations between Mendelian and complex diseases.
Drivas, T. G., Wojno, A. P., Tucker, B. A., Stone, E. M. & Bennett, J. Basal exon skipping and genetic pleiotropy: A predictive model of disease pathogenesis. Sci. Transl. Med. 10, 291ra97 (2015).
Dickey, T. H., Altschuler, S. E. & Wuttke, D. S. Single-stranded DNA-binding proteins: multiple domains for multiple functions. Structure 21, 1074–1084 (2013).
Bossi, A. & Lehner, B. Tissue specificity and the human protein interaction network. Mol. Syst. Biol. 5, 260 (2009).
Verstraeten, A., Alaerts, M., Van Laer, L. & Loeys, B. Marfan syndrome and related disorders: 25 years of gene discovery. Hum. Mutat. 37, 524–531 (2016).
Lomas, D. A. Does protease-antiprotease imbalance explain chronic obstructive pulmonary disease? Ann. Am. Thorac. Soc. 13, S130–S137 (2016).
Wong, T. Y., Cheung, C. M. G., Larsen, M., Sharma, S. & Simó, R. Diabetic retinopathy. Nat. Rev. Dis. Prim. 2, 16012 (2016).
Denny, J. C. et al. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat. Biotechnol. 31, 1102–1111 (2013).
Bush W. S., Oetjens M. T., & Crawford, D. C. Unravelling the human genome-phenome relationship using phenome-wide association studies. Nat. Rev. Genet. 17, 129–145 (2016).
Rastegar-Mojarad, M., Ye, Z., Kolesar, J. M., Hebbring, S. J. & Lin, S. M. Opportunities for drug repositioning from phenome-wide association studies. Nat. Biotechnol. 33, 342–345 (2015).
Blair, D. R. et al. A nondegenerate code of deleterious variants in Mendelian loci contributes to complex disease risk. Cell 155, 70–80 (2013).
Han, B. et al. A method to decipher pleiotropy by detecting underlying heterogeneity driven by hidden subgroups applied to autoimmune and neuropsychiatric diseases. Nat. Genet. 48, 803–810 (2016).
Queitsch, C., Carlson, K. D. & Girirajan, S. Lessons from model organisms: phenotypic robustness and missing heritability in complex disease. PLoS Genet. 8, e1003041 (2012).
Kitano, H. Biological robustness. Nat. Rev. Genet. 5, 826–837 (2004).
Hsiao, T. L. & Vitkup, D. Role of duplicate genes in robustness against deleterious human mutations. PLoS Genet. 4, e1000014 (2008).
Queitsch, C., Sangster, T. A. & Lindquist, S. Hsp90 as a capacitor of phenotypic variation. Nature 417, 618–624 (2002).
Barabasi, A.-L., Oltvai, Z. N. Z. N. & Barabási, A.-L. Network biology: understanding the cell's functional organization. Nat. Rev. Genet. 5, 101–113 (2004).
Kitano, H. et al. Metabolic syndrome and robustness tradeoffs. Diabetes 53, S6–S15 (2004).
Eichler, E. E. et al. Missing heritability and strategies for finding the underlying causes of complex disease. Nat. Rev. Genet. 11, 446–450 (2010).
Shao, H. et al. Genetic architecture of complex traits: large phenotypic effects and pervasive epistasis. Proc. Natl Acad. Sci. USA 105, 19910–19914 (2008).
Zuk, O., Hechter, E., Sunyaev, S. R. & Lander, E. S. The mystery of missing heritability: genetic interactions create phantom heritability. Proc. Natl Acad. Sci. USA 109, 1193–1198 (2012).
Bloom, J. S., Ehrenreich, I. M., Loo, W. T., Lite, T.-L. V. & Kruglyak, L. Finding the sources of missing heritability in a yeast cross. Nature 494, 234–237 (2013).
Wang, Y. et al. Parameters in dynamic models of complex traits are containers of missing heritability. PLoS Comput. Biol. 8, e1002459 (2012).
Wagner, A. Causal drift, robust signaling, and complex disease. PLoS ONE 10, e0118413 (2015).
Paaby, A. B. & Rockman, M. V. Cryptic genetic variation: evolution's hidden substrate. Nat. Rev. Genet. 15, 247–258 (2014).
Chen, R. et al. Analysis of 589,306 genomes identifies individuals resilient to severe Mendelian childhood diseases. Nat. Biotechnol. 34, 531–538 (2016).
Narasimhan, V. M. et al. Health and population effects of rare gene knockouts in adult humans with related parents. Science 352, 474–477 (2016).
Harper, A. R., Nayee, S. & Topol, E. J. Protective alleles and modifier variants in human health and disease. Nat. Rev. Genet. 16, 689–701 (2015).
Shtir, C. et al. Exome-based case-control association study using extreme phenotype design reveals novel candidates with protective effect in diabetic retinopathy. Hum. Genet. 135, 193–200 (2016).
Zhuang, L. et al. The Leu72Met polymorphism of the GHRL gene prevents the development of diabetic nephropathy in Chinese patients with type 2 diabetes mellitus. Mol. Cell. Biochem. 387, 19–25 (2014).
Lapice, E. et al. The PPARγ2 Pro12Ala variant is protective against progression of nephropathy in people with type 2 diabetes. J. Transl. Med. 13, 85 (2015).
Heng, H. H. Q. Missing heritability and stochastic genome alterations. Nat. Rev. Genet. 11, 813 (2010).
Zhang, F., Gu, W., Hurles, M. E. & Lupski, J. R. Copy number variation in human health, disease, and evolution. Annu. Rev. Genom. Hum. Genet. 10, 451–481 (2009).
Girirajan, S., Campbell, C. D. & Eichler, E. E. Human copy number variation and complex genetic disease. Annu. Rev. Genet. 45, 203–226 (2011).
Zarrei, M., MacDonald, J. R., Merico, D. & Scherer, S. W. A copy number variation map of the human genome. Nat. Rev. Genet. 16, 172–183 (2015).
Mittelman, D., Sykoudis, K., Hersh, M., Lin, Y. & Wilson, J. H. Hsp90 modulates CAG repeat instability in human cells. Cell Stress Chaperones 15, 753–759 (2010).
Chen, G., Bradford, W. D., Seidel, C. W. & Li, R. Hsp90 stress potentiates rapid cellular adaptation through induction of aneuploidy. Nature 482, 246–250 (2012).
Shameer, K. et al. Translational bioinformatics in the era of real-time biomedical, health care and wellness data streams. Brief. Bioinform. http://dx.doi.org/10.1093/bib/bbv118 (2016).
Biotechnology, N. The coming era of human phenotyping. Nat. Biotechnol. 33, 567–567 (2015).
Schork, N. J. Personalized medicine: time for one-person trials. Nature 520, 609–611 (2015).
Muoio, D. M. & Newgard, C. B. Mechanisms of disease: molecular and metabolic mechanisms of insulin resistance and β-cell failure in type 2 diabetes. Nat. Rev. Mol. Cell. Biol. 9, 193–205 (2008).
Kaushik, A., Bhatia, Y., Ali, S. & Gupta, D. Gene network rewiring to study melanoma stage progression and elements essential for driving melanoma. PLoS ONE 10, e0142443 (2015).
Hou, L., Chen, M., Zhang, C. K., Cho, J. & Zhao, H. Guilt by rewiring: gene prioritization through network rewiring in genome wide association studies. Hum. Mol. Genet. 23, 2780–2790 (2014).
Zeng, T., Wang, D. C., Wang, X., Xu, F. & Chen, L. Prediction of dynamical drug sensitivity and resistance by module network rewiring-analysis based on transcriptional profiling. Drug Resist. Updat. 17, 64–76 (2014).
Shou, C. et al. Measuring the evolutionary rewiring of biological networks. PLoS Comput. Biol. 7, e1001050 (2011).
Holme, P. & Saramäki, J. Temporal networks. Phys. Rep. 519, 97–125 (2013).
Kim, J., Kim, I., Han, S. K., Bowie, J. U. & Kim, S. Network rewiring is an important mechanism of gene essentiality change. Sci. Rep. 2, 900 (2012).
Hein, M. Y. et al. A human interactome in three quantitative dimensions organized by stoichiometries and abundances. Cell 163, 712–723 (2015).
Kastritis, P. L. & Bonvin, A. M. J. J. On the binding affinity of macromolecular interactions: daring to ask why proteins interact. J. R. Soc. Interface 10, 20120835 (2013).
Kiel, C., Verschueren, E., Yang, J.-S. & Serrano, L. Integration of protein abundance and structure data reveals competition in the ErbB signaling network. Sci. Signal. 6, ra109 (2013).
Stingele, S. et al. Global analysis of genome, transcriptome and proteome reveals the response to aneuploidy in human cells. Mol. Syst. Biol. 8, 608 (2012).
Purvis, J. E. et al. p53 dynamics control cell fate. Science 336, 1440–1444 (2012).
Moni, M. A. & Liò, P. Network-based analysis of comorbidities risk during an infection: SARS & HIV case studies. BMC Bioinformatics 15, 333 (2014).
Albert, R., Jeong, H. & Barabasi, A. Error and attack tolerance of complex networks. Nature 406, 378–382 (2000).
Yu, H., Greenbaum, D., Xin Lu, H., Zhu, X. & Gerstein, M. Genomic analysis of essentiality within protein networks. Trends Genet. 20, 227–231 (2004).
Yu, H., Kim, P. M., Sprecher, E., Trifonov, V. & Gerstein, M. The importance of bottlenecks in protein networks: correlation with gene essentiality and expression dynamics. PLoS Comput. Biol. 3, e59 (2007).
Joy, M. P., Brock, A., Ingber, D. E. & Huang, S. High-betweenness proteins in the yeast protein interaction network. J. Biomed. Biotechnol. 2005, 96–103 (2005).
Hahn, M. W. & Kern, A. D. Comparative genomics of centrality and essentiality in three eukaryotic protein-interaction networks. Mol. Biol. Evol. 22, 803–806 (2005).
Cai, J. J., Borenstein, E. & Petrov, D. A. Broker genes in human disease. 2, 815–825 (2010).
Kolch, W., Halasz, M., Granovskaya, M. & Kholodenko, B. N. The dynamic control of signal transduction networks in cancer cells. Nat. Rev. Cancer 15, 515–527 (2015).
Promislow, D. E. L. Protein networks, pleiotropy and the evolution of senescence. Proc. Biol. Sci. 271, 1225–1234 (2004).
Costanzo, M. et al. The genetic landscape of a cell. Science 327, 425–431 (2010).
Levy, S. F. & Siegal, M. L. Network hubs buffer environmental variation in Saccharomyces cerevisiae. PLoS Biol. 6, 2588–2604 (2008).
Nguyen, T.-P., Liu, W. & Jordán, F. Inferring pleiotropy by network analysis: linked diseases in the human PPI network. BMC Syst. Biol. 5, 179 (2011).
He, X. & Zhang, J. Toward a molecular understanding of pleiotropy. Genetics 173, 1885–1891 (2006).
Garcia-alonso, L. et al. The role of the interactome in the maintenance of deleterious variability in human populations. Mol. Syst. Biol. 10, 752 (2014).
Azevedo, H. & Moreira-Filho, C. A. Topological robustness analysis of protein interaction networks reveals key targets for overcoming chemotherapy resistance in glioma. Sci. Rep. 5, 16830 (2015).
Gutierrez-Arcelus, M., Rich, S. S. & Raychaudhuri, S. Autoimmune diseases — connecting risk alleles with molecular traits of the immune system. Nat. Rev. Genet. 17, 160–174 (2016).
Sollid, L. M. & Jabri, B. Triggers and drivers of autoimmunity: lessons from coeliac disease. Nat. Rev. Immunol. 13, 294–302 (2013).
Costenbader, K. H., Feskanich, D., Mandl, L. A. & Karlson, E. W. Smoking intensity, duration, and cessation, and the risk of rheumatoid arthritis in women. Am. J. Med. 119, 503.e1–503.e9 (2006).
Klareskog, L. et al. A new model for an etiology of rheumatoid arthritis: smoking may trigger HLA-DR (shared epitope)-restricted immune reactions to autoantigens modified by citrullination. Arthritis Rheum. 54, 38–46 (2006).
James, J. A. et al. Systemic lupus erythematosus in adults is associated with previous Epstein-Barr virus exposure. Arthritis Rheum. 44, 1122–1126 (2001).
Criswell, L. A. et al. Analysis of families in the Multiple Autoimmune Disease Genetics Consortium (MADGC) collection: the PTPN22 620W allele associates with multiple autoimmune phenotypes. Am. J. Hum. Genet. 76, 561–571 (2005).
Park, J.-H. et al. Estimation of effect size distribution from genome-wide association studies and implications for future discoveries. Nat. Genet. 42, 570–575 (2010).
Graham, R. R. et al. Three functional variants of IFN regulatory factor 5 (IRF5) define risk and protective haplotypes for human lupus. Proc. Natl Acad. Sci. USA 104, 6758–6763 (2007).
Dendrou, C. A. et al. Cell-specific protein phenotypes for the autoimmune locus IL2RA using a genotype-selectable human bioresource. Nat. Genet. 41, 1011–1015 (2009).
Kofler, D. M., Severson, C. a, Mousissian, N. & De Jager, P. L. and Hafler, D. A. The CD6 multiple sclerosis susceptibility allele is associated with alterations in CD4+ T cell proliferation. J. Immunol. 187, 3286–3291 (2011).
Lewis, M. J. et al. UBE2L3 polymorphism amplifies NF-κB activation and promotes plasma cell development, linking linear ubiquitination to multiple autoimmune diseases. Am. J. Hum. Genet. 96, 221–234 (2015).
Fairfax, B. P. et al. Innate immune activity conditions the effect of regulatory variants upon monocyte gene expression. Science 343, 1246949 (2014).
Shi, J. et al. Emerging role and therapeutic implication of wnt signaling pathways in autoimmune diseases. J. Immunol. Res. 2016, 9392132 (2016).
Ding, Y., Shen, S., Lino, A. C., Curotto de Lafaille, M. A. & Lafaille, J. J. β-catenin stabilization extends regulatory T cell survival and induces anergy in nonregulatory T cells. Nat. Med. 14, 162–169 (2008).
Suryawanshi, A. et al. Canonical wnt signaling in dendritic cells regulates Th1/Th17 responses and suppresses autoimmune neuroinflammation. J. Immunol. 194, 3295–3304 (2015).
Rabelo, F. d S. et al. The Wnt signaling pathway and rheumatoid arthritis. Autoimmun. Rev. 9, 207–210 (2010).
Biros, I. & Forrest, S. Spinal muscular atrophy: untangling the knot? J. Med. Genet. 36, 1–8 (1999).
Feldkötter, M., Schwarzer, V., Wirth, R., Wienker, T. F. & Wirth, B. Quantitative analyses of SMN1 and SMN2 based on real-time lightCycler PCR: fast and highly reliable carrier testing and prediction of severity of spinal muscular atrophy. Am. J. Hum. Genet. 70, 358–368 (2002).
Mailman, M. D. et al. Molecular analysis of spinal muscular atrophy and modification of the phenotype by SMN2. Genet. Med. 4, 20–26 (2002).
Lillie, E. O. et al. The n-of-1 clinical trial: the ultimate strategy for individualizing medicine? Per. Med. 8, 161–173 (2011).
Liu, J. et al. Comorbidity analysis according to sex and age in hypertension patients in china. Int. J. Med. Sci. 13, 99–107 (2016).
Vogel, F. A preliminary estimate of the number of human genes. Nature 201, 847 (1964).
US Department of Health and Human Services. Understanding our genetic inheritance, The U.S. Human Genome Project: The first five years: fiscal years 1991–1995. (US Dept. of Energy,1990).
Pertea, M. & Salzberg, S. L. Between a chicken and a grape: estimating the number of human genes. Genome Biol. 11, 206 (2010).
Menche, J. Uncovering disease-disease relationships through the incomplete interactome. Science 347, 1257601 (2015).
Gstaiger, M. & Aebersold, R. Applying mass spectrometry-based proteomics to genetics, genomics and network biology. Nat. Rev. Genet. 10, 617–627 (2009).
Collins, B. C. et al. Quantifying protein interaction dynamics by SWATH mass spectrometry: application to the 14-3-3 system. Nat. Methods 10, 1246–1253 (2013).
Humphrey, S. J., Azimifar, S. B. & Mann, M. High-throughput phosphoproteomics reveals in vivo insulin signaling dynamics. Nat. Biotechnol. 33, 990–995 (2015).
Daub, H. et al. Kinase-selective enrichment enables quantitative phosphoproteomics of the kinome across the cell cycle. Mol. Cell 31, 438–448 (2008).
Chattopadhyay, P. K., Gierahn, T. M., Roederer, M. & Love, J. C. Single-cell technologies for monitoring immune systems. Nat. Immunol. 15, 128–135 (2014).
Barabasi, A. L. & Albert, R. Emergence of scaling in random networks. Science 286, 509–512 (1999).
Mitra, K., Carvunis, A.-R., Ramesh, S. K. & Ideker, T. Integrative approaches for finding modular structure in biological networks. Nat. Rev. Genet. 14, 719–732 (2013).
Kitano, H. A robustness-based approach to systems-oriented drug design. Nat. Rev. Drug Discov. 6, 202–210 (2007).
Baffy, G. & Loscalzo, J. Complexity and network dynamics in physiological adaptation: an integrated view. Physiol. Behav. 131, 49–56 (2014).
de Lichtenberg, U. Dynamic complex formation during the yeast cell cycle. Science 307, 724–727 (2005).
Faisal, F. E. & Milenkovi, T. Dynamic networks reveal key players in aging. Bioinformatics 30, 1721–1729 (2014).
Luscombe, N. M. et al. Genomic analysis of regulatory network dynamics reveals large topological changes. Nature 431, 308–312 (2004).
Roguev, A. et al. Conservation and rewiring of functional modules revealed by an epistasis map in fission yeast. Science 322, 405–410 (2008).
Altelaar, a F. M., Munoz, J. & Heck, A. J. R. Next-generation proteomics: towards an integrative view of proteome dynamics. Nat. Rev. Genet. 14, 35–48 (2013).
Barrios-Rodiles, M. et al. High-throughput mapping of a dynamic signaling network in mammalian cells. Science 307, 1621–1625 (2005).
Dalianis, H., Hassel, M. & Velupillai, S. The Stockholm EPR corpus: characteristics and some initial findings. 14th Int. Symp. Health Inf. Manag. Res. 219, 243–249 (2009).
McGeachie, M. J. et al. Joint GWAS analysis: comparing similar GWAS at different genomic resolutions identifies novel pathway associations with six complex diseases. Genom. Data 2, 202–211 (2014).
Pendergrass, S. a et al. Phenome-wide association study (PheWAS) for detection of pleiotropy within the Population Architecture using Genomics and Epidemiology (PAGE) Network. PLoS Genet. 9, e1003087 (2013).
Glymour, M. M., Tchetgen Tchetgen, E. J. & Robins, J. M. Credible Mendelian randomization studies: approaches for evaluating the instrumental variable assumptions. Am. J. Epidemiol. 175, 332–339 (2012).
Lawlor, D. A., Harbord, R. M., Sterne, J. A. C., Timpson, N. & Davey Smith, G. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat. Med. 27, 1133–1163 (2008).
Assenov, Y., Ramírez, F., Schelhorn, S.-E., Lengauer, T. & Albrecht, M. Computing topological parameters of biological networks. Bioinformatics 24, 282–284 (2008).
Li, F. et al. PerturbationAnalyzer: a tool for investigating the effects of concentration perturbation on protein interaction networks. Bioinformatics 26, 275–277 (2010).
Zhang, B. et al. DDN: a caBIG® analytical tool for differential network analysis. Bioinformatics 27, 1036–1038 (2011).
Acknowledgements
The authors thank L. J. Jensen, C. Workman and H. V. Cook for comments on the manuscript, and D. Westergaard, J. M. Gonzalez-Izarzugaza and K. Banasik for useful discussions and suggestions. The work was supported by the Novo Nordisk Foundation (grant agreement NNF14CC0001), as well as the Innovation Fund Denmark.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Glossary
- Pleiotropy
-
The property of a genetic locus that affects more than one trait.
- Robustness
-
The property that allows a system to maintain its functions against internal and external perturbations.
- Rewiring
-
Restructuring of interactions between biological components due to conditional changes.
- Complex disease
-
A disease that is a result of complex interactions between genetics and environment that is hard to explain by a few factors.
- Multifunctionalities
-
Properties of a biological component that have multiple distinct roles.
- Comorbidities
-
Diseases that co-occur on top of a primary disease of interest in an individual.
- Genetic interaction networks
-
Networks in which nodes are genes and edges are their epistatic interactions.
- Physical interaction networks
-
Networks in which nodes physically interact. In biology interactions may be between and among, for example, proteins, DNA and RNA.
- Differential networks
-
Analytical approaches to identify edge changes between two static network states.
- Hub
-
A hub node in a network has a high degree of edges, meaning that it interacts with many other nodes in the network.
- Organizational levels
-
Levels in the hierarchy of biological structures and systems such as protein, cell, tissue, organ or organism.
- Dynamic network
-
A network that continuously changes topology over time.
- Multimorbidity
-
The coexistence of two or more diseases in the same individual without disease prioritization.
- Health transaction data
-
Data describing patients' contacts with the health care system. Data accumulates in electronic patient records and registries.
- Inversely comorbid
-
Diseases that co-occur less often in an individual than expected given their individual frequencies in the population.
- Drug repurposing
-
The application of a known drug to new indications. Synonymous with the term drug repositioning.
- Scale-free
-
A network structure that has a degree distribution following a power law.
- Bow tie
-
A multi-layered network structure where intermediate layers have far fewer components than input and output layers.
- Modularity
-
A network structure with dense connections between clusters of nodes and sparse connections between nodes in different clusters.
- Homeostasis
-
The ability to sustain various physiological parameters in a steady state.
- Plasticity
-
Variation of a phenotype as a response to a given environmental exposure.
- Epistasis
-
A phenomenon in which the function of one gene affects the function of another gene in a non-additive manner.
- Penetrance
-
From a genome-wide association study perspective, penetrance describes the proportion of individuals for which a genetic variant results in a changed phenotype.
- Network topology
-
The layout of nodes and edges in a network.
- Microsatellites
-
Polymorphic DNA loci containing repeated nucleotide sequences of typically 2–7 nucleotides per unit.
- Cryptic variation
-
Genetic variation that has little or no effect on phenotypic variation under normal conditions, but can generate heritable phenotypic variation when circumstances change.
- Edge
-
An edge represents the interaction between nodes in a network. In biological systems an edge can represent a physical interaction between two proteins or the co-occurrence of two diseases.
- Wearables
-
Personal portable devices that monitor the state of an individual.
- Nodes
-
In biological networks nodes are connection points, for example, of proteins, genes or diseases. They may or may not directly interact.
- Bottleneck
-
A bottleneck node in a network has a high degree of intersections (high betweenness), meaning that it will often be a linker between different subnetworks.
Rights and permissions
About this article
Cite this article
Hu, J., Thomas, C. & Brunak, S. Network biology concepts in complex disease comorbidities. Nat Rev Genet 17, 615–629 (2016). https://doi.org/10.1038/nrg.2016.87
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrg.2016.87
This article is cited by
-
Jacalin-related lectin 45 (OsJRL45) isolated from ‘sea rice 86’ enhances rice salt tolerance at the seedling and reproductive stages
BMC Plant Biology (2023)
-
Combination of bioaffinity ultrafiltration-UFLC-ESI-Q/TOF-MS/MS, in silico docking and multiple complex networks to explore antitumor mechanism of topoisomerase I inhibitors from Artemisiae Scopariae Herba
BMC Complementary Medicine and Therapies (2023)
-
Exploring the molecular mechanism underlying the psoriasis and T2D by using microarray data analysis
Scientific Reports (2023)
-
Enhancer promoter interactome and Mendelian randomization identify network of druggable vascular genes in coronary artery disease
Human Genomics (2022)
-
Integrative Proposals of Sports Monitoring: Subjective Outperforms Objective Monitoring
Sports Medicine - Open (2022)