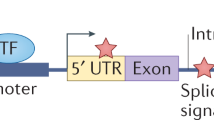
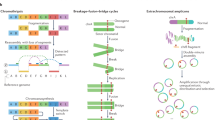
Key Points
-
Germline and somatic sequence variants in non-coding regions can play an important role in cancer.
-
Many different modes of action of non-coding variants are known. For example, point mutations and complex genomic rearrangements can disrupt or create transcription factor-binding sites or affect non-coding RNA loci.
-
Oncogenesis involves an interplay between germline and somatic variants.
-
Drivers in non-coding regions can be identified using computational methods that analyse functional effects of variants and recurrence across multiple samples.
-
Functional effects of non-coding variants can be studied by various experimental approaches.
-
The overall role of non-coding variants in tumorigenesis is currently likely underestimated as only a handful of genome-wide studies of tumours have analysed them. However, current and future efforts involving large-scale whole-genome sequencing of tumours are likely to shed more light on the importance of non-coding variants in cancer.
Abstract
Patients with cancer carry somatic sequence variants in their tumour in addition to the germline variants in their inherited genome. Although variants in protein-coding regions have received the most attention, numerous studies have noted the importance of non-coding variants in cancer. Moreover, the overwhelming majority of variants, both somatic and germline, occur in non-coding portions of the genome. We review the current understanding of non-coding variants in cancer, including the great diversity of the mutation types — from single nucleotide variants to large genomic rearrangements — and the wide range of mechanisms by which they affect gene expression to promote tumorigenesis, such as disrupting transcription factor-binding sites or functions of non-coding RNAs. We highlight specific case studies of somatic and germline variants, and discuss how non-coding variants can be interpreted on a large-scale through computational and experimental methods.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Kandoth, C. et al. Mutational landscape and significance across 12 major cancer types. Nature 502, 333–339 (2013).
Easton, D. F. & Eeles, R. A. Genome-wide association studies in cancer. Hum. Mol. Genet. 17, R109–R115 (2008).
Maurano, M. T. et al. Systematic localization of common disease-associated variation in regulatory DNA. Science 337, 1190–1195 (2012).
Chen, C. Y., Chang, I. S., Hsiung, C. A. & Wasserman, W. W. On the identification of potential regulatory variants within genome wide association candidate SNP sets. BMC Med. Genomics 7, 34 (2014).
Akhtar-Zaidi, B. et al. Epigenomic enhancer profiling defines a signature of colon cancer. Science 336, 736–739 (2012).
Kron, K. J., Bailey, S. D. & Lupien, M. Enhancer alterations in cancer: a source for a cell identity crisis. Genome Med. 6, 77 (2014).
Prensner, J. R. & Chinnaiyan, A. M. The emergence of lncRNAs in cancer biology. Cancer Discov. 1, 391–407 (2011).
Iyer, M. K. et al. The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 47, 199–208 (2015).
Stirzaker, C., Taberlay, P. C., Statham, A. L. & Clark, S. J. Mining cancer methylomes: prospects and challenges. Trends Genet. 30, 75–84 (2014).
The 1000 Genomes Project Consortium. An integrated map of genetic variation from 1,092 human genomes. Nature 491, 56–65 (2012).
Alexandrov, L. B. et al. Signatures of mutational processes in human cancer. Nature 500, 415–421 (2013).
Abyzov, A. et al. Somatic copy number mosaicism in human skin revealed by induced pluripotent stem cells. Nature 492, 438–442 (2012).
De, S. Somatic mosaicism in healthy human tissues. Trends Genet. 27, 217–223 (2011).
Lawrence, M. S. et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499, 214–218 (2013). Shows how mutational heterogeneity in the genome can lead to false positives during the identification of cancer driver genes.
Horn, S. et al. TERT promoter mutations in familial and sporadic melanoma. Science 339, 959–961 (2013). One of the first papers showing prevalence of TERT promoter mutations in cancer.
Daye, Z. J., Li, H. & Wei, Z. A powerful test for multiple rare variants association studies that incorporates sequencing qualities. Nucleic Acids Res. 40, e60 (2012).
Baca, S. C. et al. Punctuated evolution of prostate cancer genomes. Cell 153, 666–677 (2013).
Stephens, P. J. et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell 144, 27–40 (2011).
Holland, A. J. & Cleveland, D. W. Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis. Nat. Rev. Mol. Cell Biol. 10, 478–487 (2009).
Nik-Zainal, S. et al. Mutational processes molding the genomes of 21 breast cancers. Cell 149, 979–993 (2012).
Wittkopp, P. J. & Kalay, G. Cis-regulatory elements: molecular mechanisms and evolutionary processes underlying divergence. Nat. Rev. Genet. 13, 59–69 (2012).
Galas, D. J. & Schmitz, A. DNAse footprinting: a simple method for the detection of protein−DNA binding specificity. Nucleic Acids Res. 5, 3157–3170 (1978).
Neph, S. et al. An expansive human regulatory lexicon encoded in transcription factor footprints. Nature 489, 83–90 (2012).
Dunham, I. et al. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012). Discussion of functional annotations from the ENCODE project.
Hughes, J. R. et al. Analysis of hundreds of cis-regulatory landscapes at high resolution in a single, high-throughput experiment. Nat. Genet. 46, 205–212 (2014).
de Laat, W. & Dekker, J. 3C-based technologies to study the shape of the genome. Methods 58, 189–191 (2012).
Yip, K. Y. et al. Classification of human genomic regions based on experimentally determined binding sites of more than 100 transcription-related factors. Genome Biol. 13, R48 (2012).
The GTEx Consortium. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348, 648–660 (2015).
Gerstein, M. B. et al. Architecture of the human regulatory network derived from ENCODE data. Nature 489, 91–100 (2012).
Chadwick, L. H. The NIH Roadmap Epigenomics Program data resource. Epigenomics 4, 317–324 (2012).
Kundaje, A. et al. Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330 (2015).
Shalem, O. et al. Systematic dissection of the sequence determinants of gene 3′ end mediated expression control. PLoS Genet. 11, e1005147 (2015).
Dvir, S. et al. Deciphering the rules by which 5′-UTR sequences affect protein expression in yeast. Proc. Natl Acad. Sci. USA 110, E2792–E2801 (2013).
Lappalainen, T. et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature 501, 506–511 (2013).
The GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 45, 580–585 (2013).
Morris, K. V. & Mattick, J. S. The rise of regulatory RNA. Nat. Rev. Genet. 15, 423–437 (2014).
Guttman, M. & Rinn, J. L. Modular regulatory principles of large non-coding RNAs. Nature 482, 339–346 (2012).
Zhao, J., Sun, B. K., Erwin, J. A., Song, J. J. & Lee, J. T. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 322, 750–756 (2008).
Rinn, J. L. et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129, 1311–1323 (2007).
Penny, G. D., Kay, G. F., Sheardown, S. A., Rastan, S. & Brockdorff, N. Requirement for Xist in X chromosome inactivation. Nature 379, 131–137 (1996).
Schmitz, K. M., Mayer, C., Postepska, A. & Grummt, I. Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev. 24, 2264–2269 (2010).
Zhang, Z. et al. PseudoPipe: an automated pseudogene identification pipeline. Bioinformatics 22, 1437–1439 (2006).
Khurana, E. et al. Segmental duplications in the human genome reveal details of pseudogene formation. Nucleic Acids Res. 38, 6997–7007 (2010).
Sasidharan, R. & Gerstein, M. Genomics: protein fossils live on as RNA. Nature 453, 729–731 (2008).
Tam, O. H. et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature 453, 534–538 (2008).
Loots, G. G. et al. Identification of a coordinate regulator of interleukins 4, 13, and 5 by cross-species sequence comparisons. Science 288, 136–140 (2000).
Pennacchio, L. A. & Rubin, E. M. Genomic strategies to identify mammalian regulatory sequences. Nat. Rev. Genet. 2, 100–109 (2001).
Waterston, R. H. et al. Initial sequencing and comparative analysis of the mouse genome. Nature 420, 520–562 (2002).
Gibbs, R. A. et al. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature 428, 493–521 (2004).
Lindblad-Toh, K. et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature 438, 803–819 (2005).
Bejerano, G. et al. Ultraconserved elements in the human genome. Science 304, 1321–1325 (2004).
Peng, J. C., Shen, J. & Ran, Z. H. Transcribed ultraconserved region in human cancers. RNA Biol. 10, 1771–1777 (2013).
Calin, G. A. et al. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell 12, 215–229 (2007).
Khurana, E. et al. Integrative annotation of variants from 1092 humans: application to cancer genomics. Science 342, 1235587 (2013). One of the first methods for genome-wide identification of non-coding candidate cancer drivers.
Katzman, S. et al. Human genome ultraconserved elements are ultraselected. Science 317, 915 (2007).
Ward, L. D. & Kellis, M. Evidence of abundant purifying selection in humans for recently acquired regulatory functions. Science 337, 1675–1678 (2012).
Visel, A., Minovitsky, S., Dubchak, I. & Pennacchio, L. A. VISTA Enhancer Browser — a database of tissue-specific human enhancers. Nucleic Acids Res. 35, D88–D92 (2007).
Weinhold, N., Jacobsen, A., Schultz, N., Sander, C. & Lee, W. Genome-wide analysis of noncoding regulatory mutations in cancer. Nat. Genet. 46, 1160–1165 (2014). Analysis of hundreds of cancer whole-genomes to identify driver mutations in non-coding regions.
Fredriksson, N. J., Ny, L., Nilsson, J. A. & Larsson, E. Systematic analysis of noncoding somatic mutations and gene expression alterations across 14 tumor types. Nat. Genet. 46, 1258–1263 (2014).
Smith, K. S. et al. Signatures of accelerated somatic evolution in gene promoters in multiple cancer types. Nucleic Acids Res. 43, 5307–5317 (2015).
Melton, C., Reuter, J. A., Spacek, D. V. & Snyder, M. Recurrent somatic mutations in regulatory regions of human cancer genomes. Nat. Genet. 47, 710–716 (2015).
Katainen, R. et al. CTCF/cohesin-binding sites are frequently mutated in cancer. Nat. Genet. 47, 818–821 (2015).
Puente, X. S. et al. Non-coding recurrent mutations in chronic lymphocytic leukaemia. Nature 526, 519–524 (2015).
Treangen, T. J. & Salzberg, S. L. Repetitive DNA and next-generation sequencing: computational challenges and solutions. Nat. Rev. Genet. 13, 36–46 (2012).
Mijuškovic´, M. et al. A streamlined method for detecting structural variants in cancer genomes by short read paired-end sequencing. PLoS ONE 7, e48314 (2012).
Meyerson, M., Gabriel, S. & Getz, G. Advances in understanding cancer genomes through second-generation sequencing. Nat. Rev. Genet. 11, 685–696 (2010).
Heidenreich, B., Rachakonda, P. S., Hemminki, K. & Kumar, R. TERT promoter mutations in cancer development. Curr. Opin. Genet. Dev. 24, 30–37 (2014).
Huang, F. W. et al. Highly recurrent TERT promoter mutations in human melanoma. Science 339, 957–959 (2013). One of the first papers showing prevalence of TERT promoter mutations in cancer.
Killela, P. J. et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc. Natl Acad. Sci. USA 110, 6021–6026 (2013).
Hnisz, D. et al. Super-enhancers in the control of cell identity and disease. Cell 155, 934–947 (2013).
Mansour, M. R. et al. An oncogenic super-enhancer formed through somatic mutation of a noncoding intergenic element. Science 346, 644–648 (2014).
Tomlins, S. A. et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science 310, 1373–1377 (2005).
Yu, J. et al. An integrated network of androgen receptor, polycomb, and TMPRSS2−ERG gene fusions in prostate cancer progression. Cancer Cell 17, 443–454 (2010).
Berger, M. F. et al. The genomic complexity of primary human prostate cancer. Nature 470, 214–220 (2011).
Weischenfeldt, J. et al. Integrative genomic analyses reveal an androgen-driven somatic alteration landscape in early-onset prostate cancer. Cancer Cell 23, 159–170 (2013).
Northcott, P. A. et al. Enhancer hijacking activates GFI1 family oncogenes in medulloblastoma. Nature 511, 428–434 (2014).
Breit, T. M. et al. Site-specific deletions involving the tal-1 and sil genes are restricted to cells of the T cell receptor α/β lineage: T cell receptor δ gene deletion mechanism affects multiple genes. J. Exp. Med. 177, 965–977 (1993).
Nambiar, M., Kari, V. & Raghavan, S. C. Chromosomal translocations in cancer. Biochim. Biophys. Acta 1786, 139–152 (2008).
Gutschner, T. & Diederichs, S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 9, 703–719 (2012).
Han, Y., Liu, Y., Nie, L., Gui, Y. & Cai, Z. Inducing cell proliferation inhibition, apoptosis, and motility reduction by silencing long noncoding ribonucleic acid metastasis-associated lung adenocarcinoma transcript 1 in urothelial carcinoma of the bladder. Urology 81, 209.e1–209.e7 (2013).
Liu, P. Y. et al. Effects of a novel long noncoding RNA, lncUSMycN, on N-Myc expression and neuroblastoma progression. J. Natl Cancer Inst. 106, dju113 (2014).
Buechner, J. & Einvik, C. N-myc and noncoding RNAs in neuroblastoma. Mol. Cancer Res. 10, 1243–1253 (2012).
Lin, P. C. et al. Epigenetic repression of miR-31 disrupts androgen receptor homeostasis and contributes to prostate cancer progression. Cancer Res. 73, 1232–1244 (2013).
Poliseno, L. et al. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 465, 1033–1038 (2010).
Karreth, F. A. et al. The BRAF pseudogene functions as a competitive endogenous RNA and induces lymphoma in vivo. Cell 161, 319–332 (2015).
Bahcall, O. G. iCOGS collection provides a collaborative model. Nat. Genet. 45, 343 (2013).
MacArthur, D. G. et al. A systematic survey of loss-of-function variants in human protein-coding genes. Science 335, 823–828 (2012).
Wang, Q., Lu, Q. & Zhao, H. A review of study designs and statistical methods for genomic epidemiology studies using next generation sequencing. Front. Genet. 6, 149 (2015).
Bond, G. L. & Levine, A. J. A single nucleotide polymorphism in the p53 pathway interacts with gender, environmental stresses and tumor genetics to influence cancer in humans. Oncogene 26, 1317–1323 (2007).
Bond, G. L. et al. A single nucleotide polymorphism in the MDM2 promoter attenuates the p53 tumor suppressor pathway and accelerates tumor formation in humans. Cell 119, 591–602 (2004).
Grisanzio, C. & Freedman, M. L. Chromosome 8q24-associated cancers and MYC. Genes Cancer 1, 555–559 (2010).
Huang, Q. et al. A prostate cancer susceptibility allele at 6q22 increases RFX6 expression by modulating HOXB13 chromatin binding. Nat. Genet. 46, 126–135 (2014).
Oldridge, D. A. et al. Genetic predisposition to neuroblastoma mediated by a LMO1 super-enhancer polymorphism. Nature 528, 418–421 (2015).
Garritano, S. et al. In-silico identification and functional validation of allele-dependent AR enhancers. Oncotarget 6, 4816–4828 (2015).
Bakker, J. L. et al. A novel splice site mutation in the noncoding region of BRCA2: implications for Fanconi anemia and familial breast cancer diagnostics. Hum. Mut. 35, 442–446 (2014).
Demichelis, F. et al. Identification of functionally active, low frequency copy number variants at 15q21.3 and 12q21.31 associated with prostate cancer risk. Proc. Natl Acad. Sci. USA 109, 6686–6691 (2012).
Chen, X. et al. Targeted resequencing of the microRNAome and 3′UTRome reveals functional germline DNA variants with altered prevalence in epithelial ovarian cancer. Oncogene 34, 2125–2137 (2015).
Yang, Q. et al. Genetic variations in miR-27a gene decrease mature miR-27a level and reduce gastric cancer susceptibility. Oncogene 33, 193–202 (2014).
Chu, M. C., Selam, F. B. & Taylor, H. S. HOXA10 regulates p53 expression and matrigel invasion in human breast cancer cells. Cancer Biol. Ther. 3, 568–572 (2004).
Li, Q. et al. Integrative eQTL-based analyses reveal the biology of breast cancer risk loci. Cell 152, 633–641 (2013).
Xu, X. et al. Variants at IRX4 as prostate cancer expression quantitative trait loci. Eur. J. Hum. Genet. 22, 558–563 (2014).
Ongen, H. et al. Putative cis-regulatory drivers in colorectal cancer. Nature http://dx.doi.org/10.1038/nature13602 (2014).
Knudson, A. G. Mutation and cancer: statistical study of retinoblastoma. Proc. Natl Acad. Sci. USA 68, 820–823 (1971).
Calin, G. A. et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl Acad. Sci. USA 101, 2999–3004 (2004).
Pasic, I. et al. Recurrent focal copy-number changes and loss of heterozygosity implicate two noncoding RNAs and one tumor suppressor gene at chromosome 3q13.31 in osteosarcoma. Cancer Res. 70, 160–171 (2010).
Liu, Q. et al. LncRNA loc285194 is a p53-regulated tumor suppressor. Nucleic Acids Res. 41, 4976–4987 (2013).
Rachakonda, P. S. et al. TERT promoter mutations in bladder cancer affect patient survival and disease recurrence through modification by a common polymorphism. Proc. Natl Acad. Sci. USA 110, 17426–17431 (2013).
Gnad, F., Baucom, A., Mukhyala, K., Manning, G. & Zhang, Z. Assessment of computational methods for predicting the effects of missense mutations in human cancers. BMC Genomics 14, S7 (2013).
Lee, S. et al. Optimal unified approach for rare-variant association testing with application to small-sample case−control whole-exome sequencing studies. Am. J. Hum. Genet. 91, 224–237 (2012).
Tamborero, D. et al. Comprehensive identification of mutational cancer driver genes across 12 tumor types. Sci. Rep. 3, 2650 (2013).
Lochovsky, L., Zhang, J., Fu, Y., Khurana, E. & Gerstein, M. LARVA: an integrative framework for large-scale analysis of recurrent variants in noncoding annotations. Nucleic Acids Res. 43, 8123–8134 (2015). Method that accounts for heterogeneity in mutation rate in non-coding regions to identify regulatory driver mutations.
Ng, P. C. & Henikoff, S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 31, 3812–3814 (2003).
Adzhubei, I. A. et al. A method and server for predicting damaging missense mutations. Nat. Methods 7, 248–249 (2010).
Polak, P. et al. Cell-of-origin chromatin organization shapes the mutational landscape of cancer. Nature 518, 360–364 (2015). Shows that somatic mutation density can be predicted based on epigenomic features from the cell of origin.
Fu, Y. et al. FunSeq2: a framework for prioritizing noncoding regulatory variants in cancer. Genome Biol. 15, 480 (2014).
O'Roak, B. J. et al. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science 41, 177–181 (2012).
Konermann, S. et al. Genome-scale transcriptional activation by an engineered CRISPR−Cas9 complex. Nature 517, 583–588 (2014).
Mogno, I., Kwasnieski, J. C. & Cohen, B. A. Massively parallel synthetic promoter assays reveal the in vivo effects of binding site variants. Genome Res. 23, 1908–1915 (2013).
Arnold, C. D. et al. Genome-wide quantitative enhancer activity maps identified by STARR-seq. Science 339, 1074–1077 (2013).
Melnikov, A. et al. Systematic dissection and optimization of inducible enhancers in human cells using a massively parallel reporter assay. Nat. Biotechnol. 30, 271–277 (2012).
Shlyueva, D., Stampfel, G. & Stark, A. Transcriptional enhancers: from properties to genome-wide predictions. Nat. Rev. Genet. 15, 272–286 (2014).
Kwasnieski, J. C., Fiore, C., Chaudhari, H. G. & Cohen, B. A. High-throughput functional testing of ENCODE segmentation predictions. Genome Res. 24, 1595–1602 (2014).
Singh, G. & Cooper, T. A. Minigene reporter for identification and analysis of cis elements and trans factors affecting pre-mRNA splicing. Biotechniques 41, 177–181 (2006).
Gaildrat, P. et al. Use of splicing reporter minigene assay to evaluate the effect on splicing of unclassified genetic variants. Methods Mol. Biol. 653, 249–257 (2010).
Poulos, R. C. et al. Systematic screening of promoter regions pinpoints functional cis-regulatory mutations in a cutaneous melanoma genome. Mol. Cancer Res. 13, 1218–1226 (2015).
van de Wetering, M. et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 161, 933–945 (2015).
Boj, S. F. et al. Organoid models of human and mouse ductal pancreatic cancer. Cell 160, 324–338 (2015).
Gao, D. et al. Organoid cultures derived from patients with advanced prostate cancer. Cell 159, 176–187 (2014).
Ermann, J. & Glimcher, L. H. After GWAS: mice to the rescue? Curr. Opin. Immunol. 24, 564–570 (2012).
Seruggia, D., Fernández, A., Cantero, M., Pelczar, P. & Montoliu, L. Functional validation of mouse tyrosinase non-coding regulatory DNA elements by CRISPR−Cas9-mediated mutagenesis. Nucleic Acids Res. 43, 4855–4867 (2015).
Mou, H., Kennedy, Z., Anderson, D. G., Yin, H. & Xue, W. Precision cancer mouse models through genome editing with CRISPR−Cas9. Genome Med. 7, 53 (2015).
Vogelstein, B. et al. Cancer genome landscapes. Science 339, 1546–1558 (2013).
The Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 511, 543–550 (2014).
Guenther, C. A., Tasic, B., Luo, L., Bedell, M. A. & Kingsley, D. M. A molecular basis for classic blond hair color in Europeans. Nat. Genet. 46, 748–752 (2014).
Davoli, T. et al. Cumulative haploinsufficiency and triplosensitivity drive aneuploidy patterns and shape the cancer genome. Cell 155, 948–962 (2013).
Xue, W. et al. A cluster of cooperating tumor-suppressor gene candidates in chromosomal deletions. Proc. Natl Acad. Sci. USA 109, 8212–8217 (2012).
Wang, K. et al. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat. Genet. 46, 573–582 (2014).
Thurman, R. E. et al. The accessible chromatin landscape of the human genome. Nature 489, 75–82 (2012).
Bush, W. S. & Moore, J. H. Chapter 11: genome-wide association studies. PLoS Comput. Biol. 8, e1002822 (2012).
Chelala, C., Khan, A. & Lemoine, N. R. SNPnexus: a web database for functional annotation of newly discovered and public domain single nucleotide polymorphisms. Bioinformatics 25, 655–661 (2009).
Dayem Ullah, A. Z., Lemoine, N. R. & Chelala, C. SNPnexus: a web server for functional annotation of novel and publicly known genetic variants (2012 update). Nucleic Acids Res. 40, W65–W70 (2012).
Wang, K., Li, M. & Hakonarson, H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164 (2010).
McLaren, W. et al. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics 26, 2069–2070 (2010).
Perera, D. et al. OncoCis: annotation of cis-regulatory mutations in cancer. Genome Biol. 15, 485 (2014).
Paila, U., Chapman, B. A., Kirchner, R. & Quinlan, A. R. GEMINI: integrative exploration of genetic variation and genome annotations. PLoS Comput. Biol. 9, e1003153 (2013).
Coetzee, S. G., Rhie, S. K., Berman, B. P., Coetzee, G. A. & Noushmehr, H. FunciSNP: an R/bioconductor tool integrating functional non-coding data sets with genetic association studies to identify candidate regulatory SNPs. Nucleic Acids Res. 40, e139 (2012).
Ward, L. D. & Kellis, M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 40, D930–D934 (2012).
Li, M. J., Wang, L. Y., Xia, Z., Sham, P. C. & Wang, J. GWAS3D: detecting human regulatory variants by integrative analysis of genome-wide associations, chromosome interactions and histone modifications. Nucleic Acids Res. 41, W150–W158 (2013).
Macintyre, G., Bailey, J., Haviv, I. & Kowalczyk, A. is-rSNP: a novel technique for in silico regulatory SNP detection. Bioinformatics 26, i524–i530 (2010).
Boyle, A. P. et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 22, 1790–1797 (2012).
Lehmann, K. V. & Chen, T. Exploring functional variant discovery in non-coding regions with SInBaD. Nucleic Acids Res. 41, e7 (2013).
Kircher, M. et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 46, 310–315 (2014).
Ritchie, G. R., Dunham, I., Zeggini, E. & Flicek, P. Functional annotation of noncoding sequence variants. Nat. Methods 11, 294–296 (2014).
Gulko, B., Hubisz, M. J., Gronau, I. & Siepel, A. A method for calculating probabilities of fitness consequences for point mutations across the human genome. Nat. Genet. 47, 276–283 (2015).
Zhou, J. & Troyanskaya, O. G. Predicting effects of noncoding variants with deep learning-based sequence model. Nat. Methods 12, 931–934 (2015).
Acknowledgements
F.D. would like to acknowledge grant IG 13562 from AIRC (Associazione Italiana per la Ricerca sul Cancro).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
FURTHER INFORMATION
Glossary
- Exome sequencing
-
Sequencing the protein-coding portion of the genome using target-enrichment and high-throughput sequencing technology.
- Driver mutations
-
Sequence variants that confer growth advantage to tumour cells.
- Passenger mutations
-
Sequence variants that do not contribute to cancer growth.
- Germline variants
-
Heritable variants that are transmitted to offspring. These variants are constitutional (that is, present in all cells of the body).
- Genome-wide association studies
-
(GWASs). Studies that interrogate multiple common genetic variants along the genome in large cohorts of individuals to evaluate whether any variant is associated with a specific trait.
- Single nucleotide variants
-
DNA sequence changes at single nucleotides.
- Somatic variants
-
Variants that are not inherited from a parent and are not transmitted to offspring.
- Penetrance
-
The proportion of individuals carrying an allele (or a genotype) that also express the trait (phenotype) associated with it.
- Chromoplexy
-
(From the Greek pleko, meaning to weave, or to braid). A class of complex somatic DNA rearrangements whereby abundant DNA deletions and intra- and inter-chromosomal translocations that have originated in an interdependent way occur within a single cell cycle.
- Chromothripsis
-
(From the Greek thripsis, meaning shattering into pieces). A clustered chromosomal rearrangement in confined genomic regions that results from a single catastrophic event, usually limited to one chromosome.
- Kataegis
-
(From the Greek kataigis, meaning thunder). A phenomenon that is characterized by large clusters of mutations (hypermutation) in the genome of cancer cells. An APOBEC family enzyme might be responsible for the kataegis process.
- Cis-regulatory regions
-
Regions that regulate the expression of genes on the same DNA molecule. These include promoters, enhancers, silencers, insulators and untranslated regions.
- Enhancers
-
Distal cis-regulatory regions bound by transcription factors that activate genes by helping the recruitment of RNA polymerase to the promoters.
- Silencers
-
Distal cis-regulatory regions bound by transcription factors that repress gene expression by preventing RNA polymerase from binding to the gene promoter.
- Insulators
-
Regions that block the interaction between enhancers and promoters.
- DNase I footprinting
-
A method to detect the exact binding sites of DNA-binding proteins based on the fact that a protein bound to DNA protects it from cleavage by DNase I.
- Chromosome conformation capture
-
(3C). A biochemical method whereby the three-dimensional organization of chromatin in living cells is fixed and analysed.
- Expression quantitative trait loci
-
(eQTLs). Loci in which DNA sequence variants are related with expression levels of mRNAs.
- Endo-siRNAs
-
Endogenously produced small interfering RNAs that regulate gene expression by binding and cleaving mRNA targets or mediating heterochromatin formation.
- Negative selection
-
Selective pressure that results in the removal of deleterious alleles.
- Single nucleotide polymorphisms
-
(SNPs). Single nucleotide variants that show variability in the human population. As used in the context of this Review, they may be common (with high allele frequency) or rare (with low allele frequency).
- Oncogene
-
A gene that is often upregulated in cancer and can lead to or promote cancer growth.
- Burden tests
-
Statistical methods to test the cumulative effect of multiple variants in a genomic region.
- Positive selection
-
Directed selection that forces the allele frequency of advantageous mutations to increase.
- Minigene assays
-
Assays using a plasmid with a minimal gene fragment necessary for the gene to be expressed. It can include exons as well as introns, and it serves as a tool for evaluating splicing patterns.
- Precision medicine
-
Medical care tailored to the individual patient, usually using the patient's genomic sequence.
Rights and permissions
About this article
Cite this article
Khurana, E., Fu, Y., Chakravarty, D. et al. Role of non-coding sequence variants in cancer. Nat Rev Genet 17, 93–108 (2016). https://doi.org/10.1038/nrg.2015.17
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrg.2015.17
This article is cited by
-
Non-canonical antigens are the largest fraction of peptides presented by MHC class I in mismatch repair deficient murine colorectal cancer
Genome Medicine (2024)
-
Landscape of enhancer disruption and functional screen in melanoma cells
Genome Biology (2023)
-
Genome-wide enhancer-gene regulatory maps link causal variants to target genes underlying human cancer risk
Nature Communications (2023)
-
Mutational landscape of cancer-driver genes across human cancers
Scientific Reports (2023)
-
Identification of non-coding silencer elements and their regulation of gene expression
Nature Reviews Molecular Cell Biology (2023)