Key Points
-
Sterol regulatory-element binding proteins (SREBPs) are transcription factors that regulate the expression of genes involved in lipid synthesis and function as key nodes of convergence and divergence within global biological signalling networks involved in various physiological and pathophysiological processes
-
Distinctive physiological roles of SREBPs have been established: SREBP1a is involved in global lipid synthesis and growth; SREBP1c is involved in fatty acid synthesis and energy storage; and SREBP2 is involved in cholesterol regulation
-
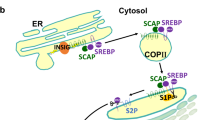
Trafficking between the endoplasmic reticulum, Golgi and nucleus are key events in the activation and regulation of SREBPs and involve factors that mediate cleavage, recycling and degradation
-
In states of energy abundance, AKT–mTOR–SREBP signalling by insulin and growth factors is the primary axis in anabolic metabolism, which produces biomass involved in nutrition, growth and cancer
-
SREBPs are involved in myriad cellular processes and pathologies such as reactive oxygen species generation, endoplasmic reticulum stress, apoptosis and autophagy; the underlying molecular mechanisms are complex and require further investigation
-
SREBP1 activation causes lipid-mediated cellular stress (lipotoxicity) that contributes to metabolic diseases such as obesity, diabetes mellitus, dyslipidaemia, hepatosteatosis and atherosclerosis, thereby further extending SREBP-related pathology to include inflammation and fibrosis in various organs
Abstract
Cellular lipid metabolism and homeostasis are controlled by sterol regulatory-element binding proteins (SREBPs). In addition to performing canonical functions in the transcriptional regulation of genes involved in the biosynthesis and uptake of lipids, genome-wide system analyses have revealed that these versatile transcription factors act as important nodes of convergence and divergence within biological signalling networks. Thus, they are involved in myriad physiological and pathophysiological processes, highlighting the importance of lipid metabolism in biology. Changes in cell metabolism and growth are reciprocally linked through SREBPs. Anabolic and growth signalling pathways branch off and connect to multiple steps of SREBP activation and form complex regulatory networks. In addition, SREBPs are implicated in numerous pathogenic processes such as endoplasmic reticulum stress, inflammation, autophagy and apoptosis, and in this way, they contribute to obesity, dyslipidaemia, diabetes mellitus, nonalcoholic fatty liver disease, nonalcoholic steatohepatitis, chronic kidney disease, neurodegenerative diseases and cancers. This Review aims to provide a comprehensive understanding of the role of SREBPs in physiology and pathophysiology at the cell, organ and organism levels.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Yokoyama, C. et al. SREBP-1, a basic-helix-loop-helix-leucine zipper protein that controls transcription of the low density lipoprotein receptor gene. Cell 75, 187–197 (1993).
Brown, M. S. & Goldstein, J. L. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell 89, 331–340 (1997).
Goldstein, J. L., DeBose-Boyd, R. A. & Brown, M. S. Protein sensors for membrane sterols. Cell 124, 35–46 (2006).
Tontonoz, P., Kim, J. B., Graves, R. A. & Spiegelman, B. M. ADD1: a novel helix-loop-helix transcription factor associated with adipocyte determination and differentiation. Mol. Cell. Biol. 13, 4753–4759 (1993).
Jeon, T. I. & Osborne, T. F. SREBPs: metabolic integrators in physiology and metabolism. Trends Endocrinol. Metab. 23, 65–72 (2012).
Shao, W. & Espenshade, P. J. Expanding roles for SREBP in metabolism. Cell Metab. 16, 414–419 (2012).
Walker, A. K. et al. A conserved SREBP-1/phosphatidylcholine feedback circuit regulates lipogenesis in metazoans. Cell 147, 840–852 (2011).
Hughes, A. L., Todd, B. L. & Espenshade, P. J. SREBP pathway responds to sterols and functions as an oxygen sensor in fission yeast. Cell 120, 831–842 (2005).
Im, S. S. et al. Linking lipid metabolism to the innate immune response in macrophages through sterol regulatory element binding protein-1a. Cell Metab. 13, 540–549 (2011).
Shimano, H. et al. Isoform 1c of sterol regulatory element binding protein is less active than isoform 1a in livers of transgenic mice and in cultured cells. J. Clin. Invest. 99, 846–854 (1997).
Toth, J. I., Datta, S., Athanikar, J. N., Freedman, L. P. & Osborne, T. F. Selective coactivator interactions in gene activation by SREBP-1a and -1c. Mol. Cell. Biol. 24, 8288–8300 (2004).
Shimano, H. et al. Overproduction of cholesterol and fatty acids causes massive liver enlargement in transgenic mice expressing truncated SREBP-1a. J. Clin. Invest. 98, 1575–1584 (1996).
Horton, J. D., Goldstein, J. L. & Brown, M. S. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Invest. 109, 1125–1131 (2002).
Amemiya-Kudo, M. et al. Transcriptional activities of nuclear SREBP-1a, -1c, and -2 to different target promoters of lipogenic and cholesterogenic genes. J. Lipid Res. 43, 1220–1235 (2002).
Oliner, J. D., Andresen, J. M., Hansen, S. K., Zhou, S. & Tjian, R. SREBP transcriptional activity is mediated through an interaction with the CREB-binding protein. Genes Dev. 10, 2903–2911 (1996).
Yang, F. et al. An ARC/Mediator subunit required for SREBP control of cholesterol and lipid homeostasis. Nature 442, 700–704 (2006).
Gong, X. et al. Structure of the WD40 domain of SCAP from fission yeast reveals the molecular basis for SREBP recognition. Cell Res. 25, 401–411 (2015).
Rawson, R. B., DeBose-Boyd, R., Goldstein, J. L. & Brown, M. S. Failure to cleave sterol regulatory element-binding proteins (SREBPs) causes cholesterol auxotrophy in Chinese hamster ovary cells with genetic absence of SREBP cleavage-activating protein. J. Biol. Chem. 274, 28549–28556 (1999).
Kuan, Y. C. et al. Heat shock protein 90 modulates lipid homeostasis by regulating the stability and function of sterol regulatory element-binding protein (SREBP) and SREBP cleavage-activating protein. J. Biol. Chem. 292, 3016–3028 (2017).
Sun, L. P., Li, L., Goldstein, J. L. & Brown, M. S. Insig required for sterol-mediated inhibition of Scap/SREBP binding to COPII proteins in vitro. J. Biol. Chem. 280, 26483–26490 (2005).
Zhang, Y. et al. Direct demonstration that loop1 of Scap binds to loop7: a crucial event in cholesterol homeostasis. J. Biol. Chem. 291, 12888–12896 (2016).
Yang, T. et al. Crucial step in cholesterol homeostasis: sterols promote binding of SCAP to INSIG-1, a membrane protein that facilitates retention of SREBPs in ER. Cell 110, 489–500 (2002).
Yabe, D., Brown, M. S. & Goldstein, J. L. Insig-2, a second endoplasmic reticulum protein that binds SCAP and blocks export of sterol regulatory element-binding proteins. Proc. Natl Acad. Sci. USA 99, 12753–12758 (2002).
Nakakuki, M. et al. A novel processing system of sterol regulatory element-binding protein-1c regulated by polyunsaturated fatty acid. J. Biochem. 155, 301–313 (2014).
Takashima, K. et al. COPI-mediated retrieval of SCAP is crucial for regulating lipogenesis under basal and sterol-deficient conditions. J. Cell Sci. 128, 2805–2815 (2015).
Xu, D. et al. PAQR3 modulates cholesterol homeostasis by anchoring Scap/SREBP complex to the Golgi apparatus. Nat. Commun. 6, 8100 (2015).
Mohn, K. L. et al. The immediate-early growth response in regenerating liver and insulin-stimulated H-35 cells: comparison with serum-stimulated 3T3 cells and identification of 41 novel immediate-early genes. Mol. Cell. Biol. 11, 381–390 (1991).
Diamond, R. H. et al. Novel delayed-early and highly insulin-induced growth response genes. Identification of HRS, a potential regulator of alternative pre-mRNA splicing. J. Biol. Chem. 268, 15185–15192 (1993).
Radhakrishnan, A., Ikeda, Y., Kwon, H. J., Brown, M. S. & Goldstein, J. L. Sterol-regulated transport of SREBPs from endoplasmic reticulum to Golgi: oxysterols block transport by binding to Insig. Proc. Natl Acad. Sci. USA 104, 6511–6518 (2007).
Ren, R. et al. Protein structure. Crystal structure of a mycobacterial Insig homolog provides insight into how these sensors monitor sterol levels. Science 349, 187–191 (2015).
Jo, Y., Lee, P. C., Sguigna, P. V. & DeBose-Boyd, R. A. Sterol-induced degradation of HMG CoA reductase depends on interplay of two Insigs and two ubiquitin ligases, gp78 and Trc8. Proc. Natl Acad. Sci. USA 108, 20503–20508 (2011).
Gong, Y. et al. Sterol-regulated ubiquitination and degradation of Insig-1 creates a convergent mechanism for feedback control of cholesterol synthesis and uptake. Cell Metab. 3, 15–24 (2006).
Liu, T. F. et al. Ablation of gp78 in liver improves hyperlipidemia and insulin resistance by inhibiting SREBP to decrease lipid biosynthesis. Cell Metab. 16, 213–225 (2012).
Yabe, D., Komuro, R., Liang, G., Goldstein, J. L. & Brown, M. S. Liver-specific mRNA for Insig-2 down-regulated by insulin: implications for fatty acid synthesis. Proc. Natl Acad. Sci. USA 100, 3155–3160 (2003).
Engelking, L. J. et al. Severe facial clefting in Insig-deficient mouse embryos caused by sterol accumulation and reversed by lovastatin. J. Clin. Invest. 116, 2356–2365 (2006).
Okada, T. et al. A serine protease inhibitor prevents endoplasmic reticulum stress-induced cleavage but not transport of the membrane-bound transcription factor ATF6. J. Biol. Chem. 278, 31024–31032 (2003).
Ye, J., Dave, U. P., Grishin, N. V., Goldstein, J. L. & Brown, M. S. Asparagine-proline sequence within membrane-spanning segment of SREBP triggers intramembrane cleavage by site-2 protease. Proc. Natl Acad. Sci. USA 97, 5123–5128 (2000).
Ye, J. et al. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol. Cell 6, 1355–1364 (2000).
Hannah, V. C., Ou, J., Luong, A., Goldstein, J. L. & Brown, M. S. Unsaturated fatty acids down-regulate srebp isoforms 1a and 1c by two mechanisms in HEK-293 cells. J. Biol. Chem. 276, 4365–4372 (2001).
Lee, J. N., Zhang, X., Feramisco, J. D., Gong, Y. & Ye, J. Unsaturated fatty acids inhibit proteasomal degradation of Insig-1 at a postubiquitination step. J. Biol. Chem. 283, 33772–33783 (2008).
Lee, J. N. et al. Identification of Ubxd8 protein as a sensor for unsaturated fatty acids and regulator of triglyceride synthesis. Proc. Natl Acad. Sci. USA 107, 21424–21429 (2010).
Morioka, S. et al. TAK1 regulates hepatic lipid homeostasis through SREBP. Oncogene 35, 3829–3838 (2016).
Aylon, Y. et al. The LATS2 tumor suppressor inhibits SREBP and suppresses hepatic cholesterol accumulation. Genes Dev. 30, 786–797 (2016).
Suchanek, M., Radzikowska, A. & Thiele, C. Photo-leucine and photo-methionine allow identification of protein–protein interactions in living cells. Nat. Methods 2, 261–267 (2005).
Wang, Q. et al. The E3 ubiquitin ligase AMFR and INSIG1 bridge the activation of TBK1 kinase by modifying the adaptor STING. Immunity 41, 919–933 (2014).
Cai, H. L. et al. A potential mechanism underlying atypical antipsychotics-induced lipid disturbances. Transl Psychiatry 5, e661 (2015).
Kabe, Y. et al. Haem-dependent dimerization of PGRMC1/Sigma-2 receptor facilitates cancer proliferation and chemoresistance. Nat. Commun. 7, 11030 (2016).
Hughes, A. L. et al. Dap1/PGRMC1 binds and regulates cytochrome P450 enzymes. Cell Metab. 5, 143–149 (2007).
Chen, Y. et al. Interferon-inducible cholesterol-25-hydroxylase inhibits hepatitis C virus replication via distinct mechanisms. Sci. Rep. 4, 7242 (2014).
Civra, A. et al. Inhibition of pathogenic non-enveloped viruses by 25-hydroxycholesterol and 27-hydroxycholesterol. Sci. Rep. 4, 7487 (2014).
Cyster, J. G., Dang, E. V., Reboldi, A. & Yi, T. 25-hydroxycholesterols in innate and adaptive immunity. Nat. Rev. Immunol. 14, 731–743 (2014).
Reboldi, A. et al. Inflammation. 25-hydroxycholesterol suppresses interleukin-1-driven inflammation downstream of type I interferon. Science 345, 679–684 (2014).
Bowie, A. The STING in the tail for cytosolic DNA-dependent activation of IRF3. Sci. Signal. 5, pe9 (2012).
Chen, W. et al. ER adaptor SCAP translocates and recruits IRF3 to perinuclear microsome induced by cytosolic microbial DNAs. PLoS Pathog. 12, e1005462 (2016).
Honda, A. et al. Cholesterol 25-hydroxylation activity of CYP3A. J. Lipid Res. 52, 1509–1516 (2011).
Hashimoto, M. et al. Knockout of mouse Cyp3a gene enhances synthesis of cholesterol and bile acid in the liver. J. Lipid Res. 54, 2060–2068 (2013).
Nagoshi, E., Imamoto, N., Sato, R. & Yoneda, Y. Nuclear import of sterol regulatory element-binding protein-2, a basic helix-loop-helix-leucine zipper (bHLH-Zip)-containing transcription factor, occurs through the direct interaction of importin β with HLH-Zip. Mol. Biol. Cell 10, 2221–2233 (1999).
Nagoshi, E. & Yoneda, Y. Dimerization of sterol regulatory element-binding protein 2 via the helix-loop-helix-leucine zipper domain is a prerequisite for its nuclear localization mediated by importin β. Mol. Cell. Biol. 21, 2779–2789 (2001).
Lee, S. J. et al. The structure of importin-beta bound to SREBP-2: nuclear import of a transcription factor. Science 302, 1571–1575 (2003).
Hirano, Y., Yoshida, M., Shimizu, M. & Sato, R. Direct demonstration of rapid degradation of nuclear sterol regulatory element-binding proteins by the ubiquitin–proteasome pathway. J. Biol. Chem. 276, 36431–36437 (2001).
Punga, T., Bengoechea-Alonso, M. T. & Ericsson, J. Phosphorylation and ubiquitination of the transcription factor sterol regulatory element-binding protein-1 in response to DNA binding. J. Biol. Chem. 281, 25278–25286 (2006).
Sundqvist, A. et al. Control of lipid metabolism by phosphorylation-dependent degradation of the SREBP family of transcription factors by SCF (Fbw7). Cell Metab. 1, 379–391 (2005).
Giandomenico, V., Simonsson, M., Gronroos, E. & Ericsson, J. Coactivator-dependent acetylation stabilizes members of the SREBP family of transcription factors. Mol. Cell. Biol. 23, 2587–2599 (2003).
Sundqvist, A. & Ericsson, J. Transcription-dependent degradation controls the stability of the SREBP family of transcription factors. Proc. Natl Acad. Sci. USA 100, 13833–13838 (2003).
Dong, Q. et al. Glycogen synthase kinase-3-mediated phosphorylation of serine 73 targets sterol response element binding protein-1c (SREBP-1c) for proteasomal degradation. Biosci. Rep. 36, e00284 (2016).
Xiong, S., Chirala, S. S. & Wakil, S. J. Sterol regulation of human fatty acid synthase promoter I requires nuclear factor-Y- and Sp-1-binding sites. Proc. Natl Acad. Sci. USA 97, 3948–3953 (2000).
Inoue, J., Sato, R. & Maeda, M. Multiple DNA elements for sterol regulatory element-binding protein and NF-Y are responsible for sterol-regulated transcription of the genes for human 3-hydroxy-3-methylglutaryl coenzyme A synthase and squalene synthase. J. Biochem. 123, 1191–1198 (1998).
Misawa, K. et al. Sterol regulatory element-binding protein-2 interacts with hepatocyte nuclear factor-4 to enhance sterol isomerase gene expression in hepatocytes. J. Biol. Chem. 278, 36176–36182 (2003).
Kanayama, T. et al. Interaction between sterol regulatory element-binding proteins and liver receptor homolog-1 reciprocally suppresses their transcriptional activities. J. Biol. Chem. 282, 10290–10298 (2007).
Louet, J. F., Hayhurst, G., Gonzalez, F. J., Girard, J. & Decaux, J. F. The coactivator PGC-1 is involved in the regulation of the liver carnitine palmitoyltransferase I gene expression by cAMP in combination with HNF4 α and cAMP-response element-binding protein (CREB). J. Biol. Chem. 277, 37991–38000 (2002).
Yamamoto, T. et al. SREBP-1 interacts with hepatocyte nuclear factor-4α and interferes with PGC-1 recruitment to suppress hepatic gluconeogenic genes. J. Biol. Chem. 279, 12027–12035 (2004).
Ponugoti, B., Fang, S. & Kemper, J. K. Functional interaction of hepatic nuclear factor-4 and peroxisome proliferator-activated receptor-γ coactivator 1α in CYP7A1 regulation is inhibited by a key lipogenic activator, sterol regulatory element-binding protein-1c. Mol. Endocrinol. 21, 2698–2712 (2007).
Lin, J. et al. Hyperlipidemic effects of dietary saturated fats mediated through PGC-1β coactivation of SREBP. Cell 120, 261–273 (2005).
Zeng, L. et al. ATF6 modulates SREBP2-mediated lipogenesis. EMBO J. 23, 950–958 (2004).
Amemiya-Kudo, M. et al. Promoter analysis of the mouse sterol regulatory element-binding protein-1c gene. J. Biol. Chem. 275, 31078–31085 (2000).
Repa, J. J. et al. Regulation of mouse sterol regulatory element-binding protein-1c gene (SREBP-1c) by oxysterol receptors, LXRα and LXRβ. Genes Dev. 14, 2819–2830 (2000).
Yoshikawa, T. et al. Identification of liver X receptor-retinoid X receptor as an activator of the sterol regulatory element-binding protein 1c gene promoter. Mol. Cell. Biol. 21, 2991–3000 (2001).
Okazaki, H., Goldstein, J. L., Brown, M. S. & Liang, G. LXR–SREBP-1c–phospholipid transfer protein axis controls very low density lipoprotein (VLDL) particle size. J. Biol. Chem. 285, 6801–6810 (2010).
Schultz, J. R. et al. Role of LXRs in control of lipogenesis. Genes Dev. 14, 2831–2838 (2000).
Kim, G. H. et al. Hepatic TRAP80 selectively regulates lipogenic activity of liver X receptor. J. Clin. Invest. 125, 183–193 (2015).
Takeuchi, Y. et al. KLF15 enables rapid switching between lipogenesis and gluconeogenesis during fasting. Cell Rep. 16, 2373–2386 (2016).
Chen, G., Liang, G., Ou, J., Goldstein, J. L. & Brown, M. S. Central role for liver X receptor in insulin-mediated activation of Srebp-1c transcription and stimulation of fatty acid synthesis in liver. Proc. Natl Acad. Sci. USA 101, 11245–11250 (2004).
Bindesboll, C. et al. Liver X receptor regulates hepatic nuclear O-GlcNAc signaling and carbohydrate responsive element-binding protein activity. J. Lipid Res. 56, 771–785 (2015).
Tian, J., Goldstein, J. L. & Brown, M. S. Insulin induction of SREBP-1c in rodent liver requires LXRα–C/EBPβ complex. Proc. Natl Acad. Sci. USA 113, 8182–8187 (2016).
Wang, G. X. et al. The brown fat-enriched secreted factor Nrg4 preserves metabolic homeostasis through attenuation of hepatic lipogenesis. Nat. Med. 20, 1436–1443 (2014).
Shimomura, I. et al. Insulin selectively increases SREBP-1c mRNA in the livers of rats with streptozotocin-induced diabetes. Proc. Natl Acad. Sci. USA 96, 13656–13661 (1999).
Wang, Y., Viscarra, J., Kim, S. J. & Sul, H. S. Transcriptional regulation of hepatic lipogenesis. Nat. Rev. Mol. Cell Biol. 16, 678–689 (2015).
Krycer, J. R., Sharpe, L. J., Luu, W. & Brown, A. J. The Akt–SREBP nexus: cell signaling meets lipid metabolism. Trends Endocrinol. Metab. 21, 268–276 (2010).
Howell, J. J., Ricoult, S. J., Ben-Sahra, I. & Manning, B. D. A growing role for mTOR in promoting anabolic metabolism. Biochem. Soc. Trans. 41, 906–912 (2013).
Zoncu, R., Efeyan, A. & Sabatini, D. M. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat. Rev. Mol. Cell Biol. 12, 21–35 (2011).
Porstmann, T. et al. SREBP activity is regulated by mTORC1 and contributes to Akt-dependent cell growth. Cell Metab. 8, 224–236 (2008).
Hagiwara, A. et al. Hepatic mTORC2 activates glycolysis and lipogenesis through Akt, glucokinase, and SREBP1c. Cell Metab. 15, 725–738 (2012).
Yecies, J. L. et al. Akt stimulates hepatic SREBP1c and lipogenesis through parallel mTORC1-dependent and independent pathways. Cell Metab. 14, 21–32 (2011).
Peterson, T. R. et al. mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell 146, 408–420 (2011).
Han, J. et al. The CREB coactivator CRTC2 controls hepatic lipid metabolism by regulating SREBP1. Nature 524, 243–246 (2015).
Duvel, K. et al. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol. Cell 39, 171–183 (2010).
Brown, M. S. & Goldstein, J. L. Selective versus total insulin resistance: a pathogenic paradox. Cell Metab. 7, 95–96 (2008).
Leavens, K. F., Easton, R. M., Shulman, G. I., Previs, S. F. & Birnbaum, M. J. Akt2 is required for hepatic lipid accumulation in models of insulin resistance. Cell Metab. 10, 405–418 (2009).
Biddinger, S. B. et al. Hepatic insulin resistance is sufficient to produce dyslipidemia and susceptibility to atherosclerosis. Cell Metab. 7, 125–134 (2008).
Li, S., Brown, M. S. & Goldstein, J. L. Bifurcation of insulin signaling pathway in rat liver: mTORC1 required for stimulation of lipogenesis, but not inhibition of gluconeogenesis. Proc. Natl Acad. Sci. USA 107, 3441–3446 (2010).
Ai, D. et al. Activation of ER stress and mTORC1 suppresses hepatic sortilin-1 levels in obese mice. J. Clin. Invest. 122, 1677–1687 (2012).
Owen, J. L. et al. Insulin stimulation of SREBP-1c processing in transgenic rat hepatocytes requires p70 S6-kinase. Proc. Natl Acad. Sci. USA 109, 16184–16189 (2012).
Khamzina, L., Veilleux, A., Bergeron, S. & Marette, A. Increased activation of the mammalian target of rapamycin pathway in liver and skeletal muscle of obese rats: possible involvement in obesity-linked insulin resistance. Endocrinology 146, 1473–1481 (2005).
Tzatsos, A. & Kandror, K. V. Nutrients suppress phosphatidylinositol 3-kinase/Akt signaling via raptor-dependent mTOR-mediated insulin receptor substrate 1 phosphorylation. Mol. Cell. Biol. 26, 63–76 (2006).
Tremblay, F. et al. Identification of IRS-1 Ser-1101 as a target of S6K1 in nutrient- and obesity-induced insulin resistance. Proc. Natl Acad. Sci. USA 104, 14056–14061 (2007).
Hsu, P. P. et al. The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science 332, 1317–1322 (2011).
Huang, J., Dibble, C. C., Matsuzaki, M. & Manning, B. D. The TSC1–TSC2 complex is required for proper activation of mTOR complex 2. Mol. Cell. Biol. 28, 4104–4115 (2008).
Yuan, M., Pino, E., Wu, L., Kacergis, M. & Soukas, A. A. Identification of Akt-independent regulation of hepatic lipogenesis by mammalian target of rapamycin (mTOR) complex 2. J. Biol. Chem. 287, 29579–29588 (2012).
Wan, M. et al. Postprandial hepatic lipid metabolism requires signaling through Akt2 independent of the transcription factors FoxA2, FoxO1, and SREBP1c. Cell Metab. 14, 516–527 (2011).
Kenerson, H. L., Subramanian, S., McIntyre, R., Kazami, M. & Yeung, R. S. Livers with constitutive mTORC1 activity resist steatosis independent of feedback suppression of Akt. PLoS ONE 10, e0117000 (2015).
Jump, D. B., Tripathy, S. & Depner, C. M. Fatty acid-regulated transcription factors in the liver. Annu. Rev. Nutr. 33, 249–269 (2013).
Tripathy, S. & Jump, D. B. Elovl5 regulates the mTORC2–Akt–FOXO1 pathway by controlling hepatic cis-vaccenic acid synthesis in diet-induced obese mice. J. Lipid Res. 54, 71–84 (2013).
Tong, X. et al. E4BP4 is an insulin-induced stabilizer of nuclear SREBP-1c and promotes SREBP-1c-mediated lipogenesis. J. Lipid Res. 57, 1219–1230 (2016).
Ide, T. et al. SREBPs suppress IRS-2-mediated insulin signalling in the liver. Nat. Cell Biol. 6, 351–357 (2004).
Kubota, N. et al. Differential hepatic distribution of insulin receptor substrates causes selective insulin resistance in diabetes and obesity. Nat. Commun. 7, 12977 (2016).
Matsuzaka, T. et al. Insulin-independent induction of sterol regulatory element-binding protein-1c expression in the livers of streptozotocin-treated mice. Diabetes 53, 560–569 (2004).
Haas, J. T. et al. Hepatic insulin signaling is required for obesity-dependent expression of SREBP-1c mRNA but not for feeding-dependent expression. Cell Metab. 15, 873–884 (2012).
Samuel, V. T. & Shulman, G. I. The pathogenesis of insulin resistance: integrating signaling pathways and substrate flux. J. Clin. Invest. 126, 12–22 (2016).
Matsumoto, M. et al. PKCλ in liver mediates insulin-induced SREBP-1c expression and determines both hepatic lipid content and overall insulin sensitivity. J. Clin. Invest. 112, 935–944 (2003).
Taniguchi, C. M. et al. Divergent regulation of hepatic glucose and lipid metabolism by phosphoinositide 3-kinase via Akt and PKCλ/ζ. Cell Metab. 3, 343–353 (2006).
Yamamoto, T. et al. Protein kinase Cβ mediates hepatic induction of sterol-regulatory element binding protein-1c by insulin. J. Lipid Res. 51, 1859–1870 (2010).
Ueki, K., Kondo, T., Tseng, Y. H. & Kahn, C. R. Central role of suppressors of cytokine signaling proteins in hepatic steatosis, insulin resistance, and the metabolic syndrome in the mouse. Proc. Natl Acad. Sci. USA 101, 10422–10427 (2004).
Sajan, M. P. et al. The critical role of atypical protein kinase C in activating hepatic SREBP-1c and NFκB in obesity. J. Lipid Res. 50, 1133–1145 (2009).
Li, Y. et al. AMPK phosphorylates and inhibits SREBP activity to attenuate hepatic steatosis and atherosclerosis in diet-induced insulin-resistant mice. Cell Metab. 13, 376–388 (2011).
Inoki, K., Zhu, T. & Guan, K. L. TSC2 mediates cellular energy response to control cell growth and survival. Cell 115, 577–590 (2003).
Gwinn, D. M. et al. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol. Cell 30, 214–226 (2008).
Lu, M. & Shyy, J. Y. Sterol regulatory element-binding protein 1 is negatively modulated by PKA phosphorylation. Am. J. Physiol. Cell Physiol. 290, C1477–C1486 (2006).
Yamamoto, T. et al. Protein kinase A suppresses sterol regulatory element-binding protein-1C expression via phosphorylation of liver X receptor in the liver. J. Biol. Chem. 282, 11687–11695 (2007).
Dong, Q. et al. Phosphorylation of sterol regulatory element binding protein-1a by protein kinase A (PKA) regulates transcriptional activity. Biochem. Biophys. Res. Commun. 449, 449–454 (2014).
Defour, A. et al. Sirtuin 1 regulates SREBP-1c expression in a LXR-dependent manner in skeletal muscle. PLoS ONE 7, e43490 (2012).
Ponugoti, B. et al. SIRT1 deacetylates and inhibits SREBP-1C activity in regulation of hepatic lipid metabolism. J. Biol. Chem. 285, 33959–33970 (2010).
Jin, S. H. et al. Resveratrol inhibits LXRα-dependent hepatic lipogenesis through novel antioxidant Sestrin2 gene induction. Toxicol. Appl. Pharmacol. 271, 95–105 (2013).
Worgall, T. S., Sturley, S. L., Seo, T., Osborne, T. F. & Deckelbaum, R. J. Polyunsaturated fatty acids decrease expression of promoters with sterol regulatory elements by decreasing levels of mature sterol regulatory element-binding protein. J. Biol. Chem. 273, 25537–25540 (1998).
Yahagi, N. et al. A crucial role of sterol regulatory element-binding protein-1 in the regulation of lipogenic gene expression by polyunsaturated fatty acids. J. Biol. Chem. 274, 35840–35844 (1999).
Jump, D. B. Dietary polyunsaturated fatty acids and regulation of gene transcription. Curr. Opin. Lipidol. 13, 155–164 (2002).
Takeuchi, Y. et al. Polyunsaturated fatty acids selectively suppress sterol regulatory element-binding protein-1 through proteolytic processing and autoloop regulatory circuit. J. Biol. Chem. 285, 11681–11691 (2010).
Kim, J. et al. Identification of Rbd2 as a candidate protease for sterol regulatory element binding protein (SREBP) cleavage in fission yeast. Biochem. Biophys. Res. Commun. 468, 606–610 (2015).
Hwang, J. et al. A Golgi rhomboid protease Rbd2 recruits Cdc48 to cleave yeast SREBP. EMBO J. 35, 2332–2349 (2016).
Guo, F. & Cavener, D. R. The GCN2 eIF2α kinase regulates fatty-acid homeostasis in the liver during deprivation of an essential amino acid. Cell Metab. 5, 103–114 (2007).
Broer, S. & Broer, A. Amino acid homeostasis and signalling in mammalian cells and organisms. Biochem. J. 474, 1935–1963 (2017).
Menendez, J. A. & Lupu, R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat. Rev. Cancer 7, 763–777 (2007).
Guo, D., Bell, E. H., Mischel, P. & Chakravarti, A. Targeting SREBP-1-driven lipid metabolism to treat cancer. Curr. Pharm. Des. 20, 2619–2626 (2014).
Ricoult, S. J., Yecies, J. L., Ben-Sahra, I. & Manning, B. D. Oncogenic PI3K and K-Ras stimulate de novo lipid synthesis through mTORC1 and SREBP. Oncogene 35, 1250–1260 (2016).
Freed-Pastor, W. A. et al. Mutant p53 disrupts mammary tissue architecture via the mevalonate pathway. Cell 148, 244–258 (2012).
Teresi, R. E., Planchon, S. M., Waite, K. A. & Eng, C. Regulation of the PTEN promoter by statins and SREBP. Hum. Mol. Genet. 17, 919–928 (2008).
Inoue, J. et al. Glutamine stimulates the gene expression and processing of sterol regulatory element binding proteins, thereby increasing the expression of their target genes. FEBS J. 278, 2739–2750 (2011).
Guo, D. et al. EGFR signaling through an Akt–SREBP-1-dependent, rapamycin-resistant pathway sensitizes glioblastomas to antilipogenic therapy. Sci. Signal. 2, ra82 (2009).
Haskins, J. W. et al. Neuregulin-activated ERBB4 induces the SREBP-2 cholesterol biosynthetic pathway and increases low-density lipoprotein uptake. Sci. Signal. 8, ra111 (2015).
Cheng, C. et al. Glucose-mediated N-glycosylation of SCAP is essential for SREBP-1 activation and tumor growth. Cancer Cell 28, 569–581 (2015).
Yamauchi, Y., Furukawa, K., Hamamura, K. & Furukawa, K. Positive feedback loop between PI3K–Akt–mTORC1 signaling and the lipogenic pathway boosts Akt signaling: induction of the lipogenic pathway by a melanoma antigen. Cancer Res. 71, 4989–4997 (2011).
Torres-Ayuso, P., Tello-Lafoz, M., Merida, I. & Avila-Flores, A. Diacylglycerol kinase-ζ regulates mTORC1 and lipogenic metabolism in cancer cells through SREBP-1. Oncogenesis 4, e164 (2015).
Zahra Bathaie, S., Ashrafi, M., Azizian, M. & Tamanoi, F. Mevalonate pathway and human cancers. Curr. Mol. Pharmacol. 10, 77–85 (2017).
Shamma, A. et al. Rb regulates DNA damage response and cellular senescence through E2F-dependent suppression of N-ras isoprenylation. Cancer Cell 15, 255–269 (2009).
Warburg, O. On the origin of cancer cells. Science 123, 309–314 (1956).
Gao, P. et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature 458, 762–765 (2009).
Xiao, D. et al. Myc promotes glutaminolysis in human neuroblastoma through direct activation of glutaminase 2. Oncotarget 6, 40655–40666 (2015).
Ricoult, S. J., Dibble, C. C., Asara, J. M. & Manning, B. D. SREBP regulates the expression and metabolic functions of wild-type and oncogenic IDH1. Mol. Cell. Biol. 36, 2384–2395 (2016).
Dasgupta, S. et al. Coactivator SRC-2-dependent metabolic reprogramming mediates prostate cancer survival and metastasis. J. Clin. Invest. 125, 1174–1188 (2015).
Liu, L. et al. Arginine methylation of SREBP1a via PRMT5 promotes de novo lipogenesis and tumor growth. Cancer Res. 76, 1260–1272 (2016).
Kamisuki, S. et al. A small molecule that blocks fat synthesis by inhibiting the activation of SREBP. Chem. Biol. 16, 882–892 (2009).
Tang, J. J. et al. Inhibition of SREBP by a small molecule, betulin, improves hyperlipidemia and insulin resistance and reduces atherosclerotic plaques. Cell Metab. 13, 44–56 (2011).
Nakakuki, M. et al. A transcription factor of lipid synthesis, sterol regulatory element-binding protein (SREBP)-1a causes G1 cell-cycle arrest after accumulation of cyclin-dependent kinase (cdk) inhibitors. FEBS J. 274, 4440–4452 (2007).
Inoue, N. et al. Lipid synthetic transcription factor SREBP-1a activates p21WAF1/CIP1, a universal cyclin-dependent kinase inhibitor. Mol. Cell. Biol. 25, 8938–8947 (2005).
Bengoechea-Alonso, M. T. & Ericsson, J. The phosphorylation-dependent regulation of nuclear SREBP1 during mitosis links lipid metabolism and cell growth. Cell Cycle 15, 2753–2765 (2016).
Griffiths, B. et al. Sterol regulatory element binding protein-dependent regulation of lipid synthesis supports cell survival and tumor growth. Cancer Metab. 1, 3 (2013).
Beloribi-Djefaflia, S., Vasseur, S. & Guillaumond, F. Lipid metabolic reprogramming in cancer cells. Oncogenesis 5, e189 (2016).
Williams, K. J. et al. An essential requirement for the SCAP/SREBP signaling axis to protect cancer cells from lipotoxicity. Cancer Res. 73, 2850–2862 (2013).
Muranaka, H. et al. A distinct function of the retinoblastoma protein in the control of lipid composition identified by lipidomic profiling. Oncogenesis 6, e350 (2017).
Hotamisligil, G. S. Endoplasmic reticulum stress and the inflammatory basis of metabolic disease. Cell 140, 900–917 (2010).
Rohrl, C. et al. Endoplasmic reticulum stress impairs cholesterol efflux and synthesis in hepatic cells. J. Lipid Res. 55, 94–103 (2014).
Lee, J. S. et al. Pharmacologic ER stress induces non-alcoholic steatohepatitis in an animal model. Toxicol. Lett. 211, 29–38 (2012).
Lee, J. N. & Ye, J. Proteolytic activation of sterol regulatory element-binding protein induced by cellular stress through depletion of Insig-1. J. Biol. Chem. 279, 45257–45265 (2004).
Lauressergues, E. et al. Does endoplasmic reticulum stress participate in APD-induced hepatic metabolic dysregulation? Neuropharmacology 62, 784–796 (2012).
Kammoun, H. L. et al. GRP78 expression inhibits insulin and ER stress-induced SREBP-1c activation and reduces hepatic steatosis in mice. J. Clin. Invest. 119, 1201–1215 (2009).
Li, H. et al. AMPK activation prevents excess nutrient-induced hepatic lipid accumulation by inhibiting mTORC1 signaling and endoplasmic reticulum stress response. Biochim. Biophys. Acta 1842, 1844–1854 (2014).
Fang, S. et al. The tumor autocrine motility factor receptor, gp78, is a ubiquitin protein ligase implicated in degradation from the endoplasmic reticulum. Proc. Natl Acad. Sci. USA 98, 14422–14427 (2001).
Zhang, T. et al. Gp78, an E3 ubiquitin ligase acts as a gatekeeper suppressing nonalcoholic steatohepatitis (NASH) and liver cancer. PLoS ONE 10, e0118448 (2015).
Wang, J. et al. n-3 polyunsaturated fatty acids protect against pancreatic β-cell damage due to ER stress and prevent diabetes development. Mol. Nutr. Food Res. 59, 1791–1802 (2015).
Sanchez-Alvarez, M. et al. Signaling networks converge on TORC1–SREBP activity to promote endoplasmic reticulum homeostasis. PLoS ONE 9, e101164 (2014).
Appenzeller-Herzog, C. & Hall, M. N. Bidirectional crosstalk between endoplasmic reticulum stress and mTOR signaling. Trends Cell Biol. 22, 274–282 (2012).
Seo, Y. K. et al. Genome-wide analysis of SREBP-1 binding in mouse liver chromatin reveals a preference for promoter proximal binding to a new motif. Proc. Natl Acad. Sci. USA 106, 13765–13769 (2009).
Wang, X. et al. Cleavage of sterol regulatory element binding proteins (SREBPs) by CPP32 during apoptosis. EMBO J. 15, 1012–1020 (1996).
Im, S. S. & Osborne, T. F. Protection from bacterial-toxin-induced apoptosis in macrophages requires the lipogenic transcription factor sterol regulatory element binding protein 1a. Mol. Cell. Biol. 32, 2196–2202 (2012).
Latz, E., Xiao, T. S. & Stutz, A. Activation and regulation of the inflammasomes. Nat. Rev. Immunol. 13, 397–411 (2013).
Schroder, K. & Tschopp, J. The inflammasomes. Cell 140, 821–832 (2010).
Karasawa, T. et al. Sterol regulatory element-binding protein-1 determines plasma remnant lipoproteins and accelerates atherosclerosis in low-density lipoprotein receptor-deficient mice. Arterioscler. Thromb. Vasc. Biol. 31, 1788–1795 (2011).
Lei, X. et al. Evidence of contribution of iPLA2β-mediated events during islet β-cell apoptosis due to proinflammatory cytokines suggests a role for iPLA2β in T1D development. Endocrinology 155, 3352–3364 (2014).
Chew, W. S. & Ong, W. Y. Regulation of calcium-independent phospholipase A2 expression by adrenoceptors and sterol regulatory element binding protein-potential crosstalk between sterol and glycerophospholipid mediators. Mol. Neurobiol. 53, 500–517 (2016).
Guan, M. et al. Nelfinavir induces liposarcoma apoptosis through inhibition of regulated intramembrane proteolysis of SREBP-1 and ATF6. Clin. Cancer Res. 17, 1796–1806 (2011).
O'Neill, L. A., Kishton, R. J. & Rathmell, J. A guide to immunometabolism for immunologists. Nat. Rev. Immunol. 16, 553–565 (2016).
van Kempen, T. S., Wenink, M. H., Leijten, E. F., Radstake, T. R. & Boes, M. Perception of self: distinguishing autoimmunity from autoinflammation. Nat. Rev. Rheumatol. 11, 483–492 (2015).
Oishi, Y. et al. SREBP1 contributes to resolution of pro-inflammatory TLR4 signaling by reprogramming fatty acid metabolism. Cell Metab. 25, 412–427 (2017).
Gilardi, F. et al. Genome-wide analysis of SREBP1 activity around the clock reveals its combined dependency on nutrient and circadian signals. PLoS Genet. 10, e1004155 (2014).
Eckel-Mahan, K. L. et al. Reprogramming of the circadian clock by nutritional challenge. Cell 155, 1464–1478 (2013).
Matsumoto, E. et al. Time of day and nutrients in feeding govern daily expression rhythms of the gene for sterol regulatory element-binding protein (SREBP)-1 in the mouse liver. J. Biol. Chem. 285, 33028–33036 (2010).
Masri, S. et al. Partitioning circadian transcription by SIRT6 leads to segregated control of cellular metabolism. Cell 158, 659–672 (2014).
Tao, R., Xiong, X., DePinho, R. A., Deng, C. X. & Dong, X. C. Hepatic SREBP-2 and cholesterol biosynthesis are regulated by FoxO3 and Sirt6. J. Lipid Res. 54, 2745–2753 (2013).
Le Martelot, G. et al. REV-ERBα participates in circadian SREBP signaling and bile acid homeostasis. PLoS Biol. 7, e1000181 (2009).
Cretenet, G., Le Clech, M. & Gachon, F. Circadian clock-coordinated 12 Hr period rhythmic activation of the IRE1α pathway controls lipid metabolism in mouse liver. Cell Metab. 11, 47–57 (2010).
Masri, S. et al. Lung adenocarcinoma distally rewires hepatic circadian homeostasis. Cell 165, 896–909 (2016).
Etchegaray, J. P. et al. Casein kinase 1δ regulates the pace of the mammalian circadian clock. Mol. Cell. Biol. 29, 3853–3866 (2009).
Brookheart, R. T., Lee, C. Y. & Espenshade, P. J. Casein kinase 1 regulates sterol regulatory element-binding protein (SREBP) to control sterol homeostasis. J. Biol. Chem. 289, 2725–2735 (2014).
Horton, J. D., Bashmakov, Y., Shimomura, I. & Shimano, H. Regulation of sterol regulatory element binding proteins in livers of fasted and refed mice. Proc. Natl Acad. Sci. USA 95, 5987–5992 (1998).
Shimano, H. et al. Sterol regulatory element-binding protein-1 as a key transcription factor for nutritional induction of lipogenic enzyme genes. J. Biol. Chem. 274, 35832–35839 (1999).
Yahagi, N. et al. Absence of sterol regulatory element-binding protein-1 (SREBP-1) ameliorates fatty livers but not obesity or insulin resistance in Lepob/Lepob mice. J. Biol. Chem. 277, 19353–19357 (2002).
Moon, Y. A. et al. The Scap/SREBP pathway is essential for developing diabetic fatty liver and carbohydrate-induced hypertriglyceridemia in animals. Cell Metab. 15, 240–246 (2012).
Liu, J. Ethanol and liver: recent insights into the mechanisms of ethanol-induced fatty liver. World J. Gastroenterol. 20, 14672–14685 (2014).
Osei-Hyiaman, D. et al. Hepatic CB1 receptor is required for development of diet-induced steatosis, dyslipidemia, and insulin and leptin resistance in mice. J. Clin. Invest. 118, 3160–3169 (2008).
Bose, S. K. et al. Forkhead box transcription factor regulation and lipid accumulation by hepatitis C virus. J. Virol. 88, 4195–4203 (2014).
McRae, S. et al. The hepatitis C virus-induced NLRP3 inflammasome activates the sterol regulatory element-binding protein (SREBP) and regulates lipid metabolism. J. Biol. Chem. 291, 3254–3267 (2016).
Pajvani, U. B. et al. Inhibition of Notch uncouples Akt activation from hepatic lipid accumulation by decreasing mTorc1 stability. Nat. Med. 19, 1054–1060 (2013).
Lee, Y. H. et al. Exogenous administration of DLK1 ameliorates hepatic steatosis and regulates gluconeogenesis via activation of AMPK. Int. J. Obes. (Lond.) 40, 356–365 (2016).
Nagaya, T. et al. Down-regulation of SREBP-1c is associated with the development of burned-out NASH. J. Hepatol. 53, 724–731 (2010).
Zhang, W. et al. Stat3 pathway correlates with the roles of leptin in mouse liver fibrosis and sterol regulatory element binding protein-1c expression of rat hepatic stellate cells. Int. J. Biochem. Cell Biol. 45, 736–744 (2013).
Zhai, X. et al. The β-catenin pathway contributes to the effects of leptin on SREBP-1c expression in rat hepatic stellate cells and liver fibrosis. Br. J. Pharmacol. 169, 197–212 (2013).
Van Rooyen, D. M. et al. Hepatic free cholesterol accumulates in obese, diabetic mice and causes nonalcoholic steatohepatitis. Gastroenterology 141, 1393–1403 (2011).
Tomita, K. et al. Free cholesterol accumulation in hepatic stellate cells: mechanism of liver fibrosis aggravation in nonalcoholic steatohepatitis in mice. Hepatology 59, 154–169 (2014).
Kang, Q. & Chen, A. Curcumin inhibits srebp-2 expression in activated hepatic stellate cells in vitro by reducing the activity of specificity protein-1. Endocrinology 150, 5384–5394 (2009).
Siersbaek, R., Nielsen, R. & Mandrup, S. PPARγ in adipocyte differentiation and metabolism — novel insights from genome-wide studies. FEBS Lett. 584, 3242–3249 (2010).
Payne, V. A. et al. C/EBP transcription factors regulate SREBP1c gene expression during adipogenesis. Biochem. J. 425, 215–223 (2010).
Kim, J. B., Wright, H. M., Wright, M. & Spiegelman, B. M. ADD1/SREBP1 activates PPARγ through the production of endogenous ligand. Proc. Natl Acad. Sci. USA 95, 4333–4337 (1998).
Fajas, L. et al. Regulation of peroxisome proliferator-activated receptor γ expression by adipocyte differentiation and determination factor 1/sterol regulatory element binding protein 1: implications for adipocyte differentiation and metabolism. Mol. Cell. Biol. 19, 5495–5503 (1999).
Shimano, H. et al. Elevated levels of SREBP-2 and cholesterol synthesis in livers of mice homozygous for a targeted disruption of the SREBP-1 gene. J. Clin. Invest. 100, 2115–2124 (1997).
Sekiya, M. et al. SREBP-1-independent regulation of lipogenic gene expression in adipocytes. J. Lipid Res. 48, 1581–1591 (2007).
Shimomura, I. et al. Insulin resistance and diabetes mellitus in transgenic mice expressing nuclear SREBP-1c in adipose tissue: model for congenital generalized lipodystrophy. Genes Dev. 12, 3182–3194 (1998).
Shimomura, I., Hammer, R. E., Ikemoto, S., Brown, M. S. & Goldstein, J. L. Leptin reverses insulin resistance and diabetes mellitus in mice with congenital lipodystrophy. Nature 401, 73–76 (1999).
Horton, J. D., Shimomura, I., Ikemoto, S., Bashmakov, Y. & Hammer, R. E. Overexpression of sterol regulatory element-binding protein-1a in mouse adipose tissue produces adipocyte hypertrophy, increased fatty acid secretion, and fatty liver. J. Biol. Chem. 278, 36652–36660 (2003).
Ayala-Sumuano, J. T. et al. Srebf1a is a key regulator of transcriptional control for adipogenesis. Sci. Rep. 1, 178 (2011).
Fujii, N. et al. Sterol regulatory element-binding protein-1c orchestrates metabolic remodeling of white adipose tissue by caloric restriction. Aging Cell 16, 508–517 (2017).
Takahashi, A. et al. Transgenic mice overexpressing nuclear SREBP-1c in pancreatic β-cells. Diabetes 54, 492–499 (2005).
Ishikawa, M. et al. Cholesterol accumulation and diabetes in pancreatic β-cell-specific SREBP-2 transgenic mice: a new model for lipotoxicity. J. Lipid Res. 49, 2524–2534 (2008).
Iwasaki, Y. et al. Nuclear SREBP-1a causes loss of pancreatic β-cells and impaired insulin secretion. Biochem. Biophys. Res. Commun. 378, 545–550 (2009).
Yamashita, T. et al. Role of uncoupling protein-2 up-regulation and triglyceride accumulation in impaired glucose-stimulated insulin secretion in a β-cell lipotoxicity model overexpressing sterol regulatory element-binding protein-1c. Endocrinology 145, 3566–3577 (2004).
Amemiya-Kudo, M. et al. Suppression of the pancreatic duodenal homeodomain transcription factor-1 (Pdx-1) promoter by sterol regulatory element-binding protein-1c (SREBP-1c). J. Biol. Chem. 286, 27902–27914 (2011).
Kato, T. et al. Granuphilin is activated by SREBP-1c and involved in impaired insulin secretion in diabetic mice. Cell Metab. 4, 143–154 (2006).
Choi, S. E. et al. Stimulation of lipogenesis as well as fatty acid oxidation protects against palmitate-induced INS-1 β-cell death. Endocrinology 152, 816–827 (2011).
Xu, F. et al. SIRT1 mediates the effect of GLP-1 receptor agonist exenatide on ameliorating hepatic steatosis. Diabetes 63, 3637–3646 (2014).
Ozbay, L. A. et al. Cyclosporin and tacrolimus impair insulin secretion and transcriptional regulation in INS-1E β-cells. Br. J. Pharmacol. 162, 136–146 (2011).
Talchai, C., Xuan, S., Lin, H. V., Sussel, L. & Accili, D. Pancreatic β cell dedifferentiation as a mechanism of diabetic β cell failure. Cell 150, 1223–1234 (2012).
Shepardson, N. E., Shankar, G. M. & Selkoe, D. J. Cholesterol level and statin use in Alzheimer disease: II. Review of human trials and recommendations. Arch. Neurol. 68, 1385–1392 (2011).
Shepardson, N. E., Shankar, G. M. & Selkoe, D. J. Cholesterol level and statin use in Alzheimer disease: I. Review of epidemiological and preclinical studies. Arch. Neurol. 68, 1239–1244 (2011).
Barbero-Camps, E., Fernandez, A., Martinez, L., Fernandez-Checa, J. C. & Colell, A. APP/PS1 mice overexpressing SREBP-2 exhibit combined Aβ accumulation and tau pathology underlying Alzheimer's disease. Hum. Mol. Genet. 22, 3460–3476 (2013).
Pierrot, N. et al. Amyloid precursor protein controls cholesterol turnover needed for neuronal activity. EMBO Mol. Med. 5, 608–625 (2013).
Suzuki, R., Ferris, H. A., Chee, M. J., Maratos-Flier, E. & Kahn, C. R. Reduction of the cholesterol sensor SCAP in the brains of mice causes impaired synaptic transmission and altered cognitive function. PLoS Biol. 11, e1001532 (2013).
Suzuki, R. et al. Diabetes and insulin in regulation of brain cholesterol metabolism. Cell Metab. 12, 567–579 (2010).
Valenza, M. et al. Disruption of astrocyte–neuron cholesterol cross talk affects neuronal function in Huntington's disease. Cell Death Differ. 22, 690–702 (2015).
Liu, L. et al. Glial lipid droplets and ROS induced by mitochondrial defects promote neurodegeneration. Cell 160, 177–190 (2015).
Raeder, M. B., Ferno, J., Glambek, M., Stansberg, C. & Steen, V. M. Antidepressant drugs activate SREBP and up-regulate cholesterol and fatty acid biosynthesis in human glial cells. Neurosci. Lett. 395, 185–190 (2006).
Raeder, M. B., Ferno, J., Vik-Mo, A. O. & Steen, V. M. SREBP activation by antipsychotic- and antidepressant-drugs in cultured human liver cells: relevance for metabolic side-effects? Mol. Cell. Biochem. 289, 167–173 (2006).
Norrmen, C. et al. mTORC1 controls PNS myelination along the mTORC1–RXRγ–SREBP-lipid biosynthesis axis in Schwann cells. Cell Rep. 9, 646–660 (2014).
Preitschopf, A. et al. mTORC1 is essential for early steps during Schwann cell differentiation of amniotic fluid stem cells and regulates lipogenic gene expression. PLoS ONE 9, e107004 (2014).
Le Hellard, S. et al. Polymorphisms in SREBF1 and SREBF2, two antipsychotic-activated transcription factors controlling cellular lipogenesis, are associated with schizophrenia in German and Scandinavian samples. Mol. Psychiatry 15, 463–472 (2010).
Steen, V. M. et al. Genetic evidence for a role of the SREBP transcription system and lipid biosynthesis in schizophrenia and antipsychotic treatment. Eur. Neuropsychopharmacol. 27, 589–598 (2016).
Gnudi, L. Angiopoietins and diabetic nephropathy. Diabetologia 59, 1616–1620 (2016).
Proctor, G. et al. Regulation of renal fatty acid and cholesterol metabolism, inflammation, and fibrosis in Akita and OVE26 mice with type 1 diabetes. Diabetes 55, 2502–2509 (2006).
Wang, Z. et al. Regulation of renal lipid metabolism, lipid accumulation, and glomerulosclerosis in FVBdb/db mice with type 2 diabetes. Diabetes 54, 2328–2335 (2005).
Ishigaki, N. et al. Involvement of glomerular SREBP-1c in diabetic nephropathy. Biochem. Biophys. Res. Commun. 364, 502–508 (2007).
Sun, L., Halaihel, N., Zhang, W., Rogers, T. & Levi, M. Role of sterol regulatory element-binding protein 1 in regulation of renal lipid metabolism and glomerulosclerosis in diabetes mellitus. J. Biol. Chem. 277, 18919–18927 (2002).
Chen, G. et al. SREBP-1 is a novel mediator of TGFβ1 signaling in mesangial cells. J. Mol. Cell. Biol. 6, 516–530 (2014).
An, W. et al. Cyclin Y is involved in the regulation of adipogenesis and lipid production. PLoS ONE 10, e0132721 (2015).
Sun, H., Yuan, Y. & Sun, Z. L. Cholesterol contributes to diabetic nephropathy through SCAP–SREBP-2 pathway. Int. J. Endocrinol. 2013, 592576 (2013).
Godel, M. et al. Role of mTOR in podocyte function and diabetic nephropathy in humans and mice. J. Clin. Invest. 121, 2197–2209 (2011).
D'Agati, V. D. et al. Obesity-related glomerulopathy: clinical and pathologic characteristics and pathogenesis. Nat. Rev. Nephrol. 12, 453–471 (2016).
Tominaga, T. et al. Transcriptional and translational modulation of myo-inositol oxygenase (Miox) by fatty acids: implications in renal tubular ijury induced in obesity and diabetes. J. Biol. Chem. 291, 1348–1367 (2016).
Shao, W., Machamer, C. E. & Espenshade, P. J. Fatostatin blocks ER exit of SCAP but inhibits cell growth in a SCAP-independent manner. J. Lipid Res. 57, 1564–1573 (2016).
Burr, R. et al. Mga2 transcription factor regulates an oxygen-responsive lipid homeostasis pathway in fission yeast. J. Biol. Chem. 291, 12171–12183 (2016).
Gholkar, A. A. et al. Fatostatin inhibits cancer cell proliferation by affecting mitotic microtubule spindle assembly and cell division. J. Biol. Chem. 291, 17001–17008 (2016).
Miyata, S., Inoue, J., Shimizu, M. & Sato, R. Xanthohumol improves diet-induced obesity and fatty liver by suppressing sterol regulatory element-binding protein (SREBP) activation. J. Biol. Chem. 290, 20565–20579 (2015).
Doddapattar, P. et al. Xanthohumol ameliorates atherosclerotic plaque formation, hypercholesterolemia, and hepatic steatosis in ApoE-deficient mice. Mol. Nutr. Food Res. 57, 1718–1728 (2013).
Aryal, B., Singh, A. K., Rotllan, N., Price, N. & Fernandez-Hernando, C. MicroRNAs and lipid metabolism. Curr. Opin. Lipidol. 28, 273–280 (2017).
Najafi-Shoushtari, S. H. et al. MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science 328, 1566–1569 (2010).
Horie, T. et al. MicroRNA-33 encoded by an intron of sterol regulatory element-binding protein 2 (Srebp2) regulates HDL in vivo. Proc. Natl Acad. Sci. USA 107, 17321–17326 (2010).
Horie, T. et al. MicroRNA-33 regulates sterol regulatory element-binding protein 1 expression in mice. Nat. Commun. 4, 2883 (2013).
Horie, T. et al. MicroRNA-33b knock-in mice for an intron of sterol regulatory element-binding factor 1 (Srebf1) exhibit reduced HDL-C in vivo. Sci. Rep 4, 5312 (2014).
Rottiers, V. & Naar, A. M. MicroRNAs in metabolism and metabolic disorders. Nat. Rev. Mol. Cell Biol. 13, 239–250 (2012).
Karunakaran, D. et al. Therapeutic inhibition of miR-33 promotes fatty acid oxidation but does not ameliorate metabolic dysfunction in diet-induced obesity. Arterioscler. Thromb. Vasc. Biol. 35, 2536–2543 (2015).
Herrera-Merchan, A. et al. miR-33-mediated downregulation of p53 controls hematopoietic stem cell self-renewal. Cell Cycle 9, 3277–3285 (2010).
Lin, Y. et al. MicroRNA-33b inhibits breast cancer metastasis by targeting HMGA2, SALL4 and Twist1. Sci. Rep. 5, 9995 (2015).
Jeon, T. I. et al. An SREBP-responsive microRNA operon contributes to a regulatory loop for intracellular lipid homeostasis. Cell Metab. 18, 51–61 (2013).
Yang, M. et al. Identification of miR-185 as a regulator of de novo cholesterol biosynthesis and low density lipoprotein uptake. J. Lipid Res. 55, 226–238 (2014).
Ru, P. et al. Feedback loop regulation of SCAP/SREBP-1 by miR-29 modulates EGFR signaling-driven glioblastoma growth. Cell Rep. 16, 1527–1535 (2016).
Everett, B. M., Smith, R. J. & Hiatt, W. R. Reducing LDL with PCSK9 inhibitors — the clinical benefit of lipid drugs. N. Engl. J. Med. 373, 1588–1591 (2015).
Horton, J. D., Cohen, J. C. & Hobbs, H. H. Molecular biology of PCSK9: its role in LDL metabolism. Trends Biochem. Sci. 32, 71–77 (2007).
Sabatine, M. S. et al. Efficacy and safety of evolocumab in reducing lipids and cardiovascular events. N. Engl. J. Med. 372, 1500–1509 (2015).
Ma, K. L. et al. Activation of mTOR modulates SREBP-2 to induce foam cell formation through increased retinoblastoma protein phosphorylation. Cardiovasc. Res. 100, 450–460 (2013).
Ai, D. et al. Regulation of hepatic LDL receptors by mTORC1 and PCSK9 in mice. J. Clin. Invest. 122, 1262–1270 (2012).
Liu, J. et al. Activation of mTORC1 disrupted LDL receptor pathway: a potential new mechanism for the progression of non-alcoholic fatty liver disease. Int. J. Biochem. Cell Biol. 61, 8–19 (2015).
Ecker, J. et al. Induction of fatty acid synthesis is a key requirement for phagocytic differentiation of human monocytes. Proc. Natl Acad. Sci. USA 107, 7817–7822 (2010).
Wei, X. et al. Fatty acid synthesis configures the plasma membrane for inflammation in diabetes. Nature 539, 294–298 (2016).
Acknowledgements
The authors sincerely thank all their collaborators and laboratory members for their contributions to the work discussed in this Review. The authors are aware that there are considerable pieces of work on SREBPs that could not be included in this Review owing to lack of space. The authors acknowledge Enago for reviewing the English language in this Review before submission of the manuscript.
Author information
Authors and Affiliations
Contributions
H.S. and R.S. researched data for the article, made substantial contributions to discussions about the content, wrote the article and reviewed and/or edited the manuscript before submission.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Shimano, H., Sato, R. SREBP-regulated lipid metabolism: convergent physiology — divergent pathophysiology. Nat Rev Endocrinol 13, 710–730 (2017). https://doi.org/10.1038/nrendo.2017.91
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrendo.2017.91
This article is cited by
-
Integrating network pharmacology and animal experimental validation to investigate the action mechanism of oleanolic acid in obesity
Journal of Translational Medicine (2024)
-
A concerted neuron–astrocyte program declines in ageing and schizophrenia
Nature (2024)
-
MEF2C regulates NK cell effector functions through control of lipid metabolism
Nature Immunology (2024)
-
Inhibition of 7-dehydrocholesterol reductase prevents hepatic ferroptosis under an active state of sterol synthesis
Nature Communications (2024)
-
Activation of the PERK-CHOP signaling pathway during endoplasmic reticulum stress contributes to olanzapine-induced dyslipidemia
Acta Pharmacologica Sinica (2024)