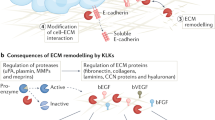
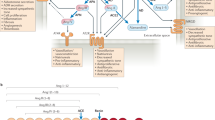
Key Points
-
Tissue kallikreins (KLKs) constitute a family of 15 secreted serine proteases that are encoded by the largest protease gene cluster in the human genome.
-
KLKs were traditionally known for their clinical applicability as cancer markers — for instance, KLK3 (also known as prostate-specific antigen) is a marker for prostate cancer.
-
The field of KLK research has recently blossomed with the development of KLK-knockout models and the elucidation of the 3D structures of several KLKs.
-
Novel pathophysiological roles for these proteases have recently been assigned in various tissues, such as the airway, cardiovascular system, tooth, skin and brain tissues.
-
The promise of KLKs as therapeutic targets in various pathologies — including skin diseases, hereditary angioedema, neurodegeneration, inflammation and cancer — is emerging.
-
Systematic efforts for the development of the first generation of KLK-based inhibitors as candidate therapeutics have already been initiated.
Abstract
Tissue kallikreins are a family of fifteen secreted serine proteases encoded by the largest protease gene cluster in the human genome. In the past decade, substantial progress has been made in characterizing the natural substrates, endogenous inhibitors and in vivo functions of kallikreins, and studies have delineated important pathophysiological roles for these proteases in a variety of tissues. Thus, kallikreins are now considered attractive targets for the development of novel therapeutics for airway, cardiovascular, tooth, brain, skin and neoplastic diseases. In this Review, we discuss recent advances in our understanding of the physiological functions and pathological implications of kallikrein proteases, and highlight progress in the identification of kallikrein inhibitors, which together are bringing us closer to therapeutically targeting kallikreins in selected disease settings.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Change history
28 August 2015
The references for compound 1 and compound 8 were incorrectly cited. The authors apologize for this unintended error, and the online version of the article has been corrected.
16 September 2015
The classes of compounds developed as KLK1 inhibitors mentioned on page 194 were modified.
References
Drag, M. & Salvesen, G. S. Emerging principles in protease-based drug discovery. Nature Rev. Drug Discov. 9, 690–701 (2010).
Borgono, C. A. & Diamandis, E. P. The emerging roles of human tissue kallikreins in cancer. Nature Rev. Cancer 4, 876–890 (2004).
Yousef, G. M., Chang, A., Scorilas, A. & Diamandis, E. P. Genomic organization of the human kallikrein gene family on chromosome 19q13.3–q13.4. Biochem. Biophys. Res. Commun. 276, 125–133 (2000).
Marceau, F. & Regoli, D. Bradykinin receptor ligands: therapeutic perspectives. Nature Rev. Drug Discov. 3, 845–852 (2004).
Lilja, H., Ulmert, D. & Vickers, A. J. Prostate-specific antigen and prostate cancer: prediction, detection and monitoring. Nature Rev. Cancer 8, 268–278 (2008).
Sotiropoulou, G. & Pampalakis, G. Kallikrein-related peptidases: bridges between immune functions and extracellular matrix degradation. Biol. Chem. 391, 321–331 (2010).
Pampalakis, G. & Sotiropoulou, G. Tissue kallikrein proteolytic cascade pathways in normal physiology and cancer. Biochim. Biophys. Acta 1776, 22–31 (2007).
Emami, N. & Diamandis, E. P. New insights into the functional mechanisms and clinical applications of the kallikrein-related peptidase family. Mol. Oncol. 1, 269–287 (2007).
Lundwall, A. & Brattsand, M. Kallikrein-related peptidases. Cell. Mol. Life Sci. 65, 2019–2038 (2008).
Goettig, P., Magdolen, V. & Brandstetter, H. Natural and synthetic inhibitors of kallikrein-related peptidases (KLKs). Biochimie 92, 1546–1567 (2010). This is a thorough review on the structural basis of KLK inhibition by endogenous inhibitors.
Sotiropoulou, G. & Pampalakis, G. Targeting the kallikrein-related peptidases for drug development. Trends Pharmacol. Sci. 33, 623–634 (2012).
Swedberg, J. E., de Veer, S. J. & Harris, J. M. Natural and engineered kallikrein inhibitors: an emerging pharmacopoeia. Biol. Chem. 391, 357–374 (2010).
Yousef, G. M. & Diamandis, E. P. The new human tissue kallikrein gene family: structure, function, and association to disease. Endocr. Rev. 22, 184–204 (2001).
Bjorkqvist, J., Jamsa, A. & Renne, T. Plasma kallikrein: the bradykinin-producing enzyme. Thromb. Haemost. 110, 399–407 (2013).
Moreau, M. E. et al. The kallikrein-kinin system: current and future pharmacological targets. J. Pharmacol. Sci. 99, 6–38 (2005).
Pathak, M., Wong, S. S., Dreveny, I. & Emsley, J. Structure of plasma and tissue kallikreins. Thromb. Haemost. 110, 423–433 (2013).
Cicardi, M. et al. Ecallantide for the treatment of acute attacks in hereditary angioedema. N. Engl. J. Med. 363, 523–531 (2010). This is the first evaluation of ecallantide in a double-blind, placebo control trial against hereditary angioedema.
Zuraw, B., Yasothan, U. & Kirkpatrick, P. Ecallantide. Nature Rev. Drug Discov. 9, 189–190 (2010).
Marcondes, S. & Antunes, E. The plasma and tissue kininogen-kallikrein-kinin system: role in the cardiovascular system. Curr. Med. Chem. Cardiovasc. Hematol. Agents 3, 33–44 (2005).
Costa-Neto, C. M. et al. Participation of kallikrein-kinin system in different pathologies. Int. Immunopharmacol. 8, 135–142 (2008).
Page, M. J. & Di Cera, E. Serine peptidases: classification, structure and function. Cell. Mol. Life Sci. 65, 1220–1236 (2008).
Rawlings, N. D., Waller, M., Barrett, A. J. & Bateman, A. MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 42, D503–D509 (2014).
Yoon, H. et al. A completed KLK activome profile: investigation of activation profiles of KLK9, 10, and 15. Biol. Chem. 390, 373–377 (2009).
Yoon, H. et al. Activation profiles and regulatory cascades of the human kallikrein-related peptidases. J. Biol. Chem. 282, 31852–31864 (2007).
Cloutier, S. M. et al. Substrate specificity of human kallikrein 2 (hK2) as determined by phage display technology. Eur. J. Biochem. 269, 2747–2754 (2002).
Coombs, G. S. et al. Substrate specificity of prostate-specific antigen (PSA). Chem. Biol. 5, 475–488 (1998).
Felber, L. M. et al. Enzymatic profiling of human kallikrein 14 using phage-display substrate technology. Biol. Chem. 386, 291–298 (2005).
Li, H. X. et al. Substrate specificity of human kallikreins 1 and 6 determined by phage display. Protein Sci. 17, 664–672 (2008).
Sharma, N. et al. Substrate specificity determination of mouse implantation serine proteinase and human kallikrein-related peptidase 6 by phage display. Biol. Chem. 389, 1097–1105 (2008).
Borgono, C. A. et al. Defining the extended substrate specificity of kallikrein 1-related peptidases. Biol. Chem. 388, 1215–1225 (2007).
de Veer, S. J., Swedberg, J. E., Parker, E. A. & Harris, J. M. Non-combinatorial library screening reveals subsite cooperativity and identifies new high-efficiency substrates for kallikrein-related peptidase 14. Biol. Chem. 393, 331–341 (2012).
Debela, M. et al. Specificity profiling of seven human tissue kallikreins reveals individual subsite preferences. J. Biol. Chem. 281, 25678–25688 (2006).
Matsumura, M. et al. Substrates of the prostate-specific serine protease prostase/KLK4 defined by positional-scanning peptide libraries. Prostate 62, 1–13 (2005).
Bourgeois, L. et al. Serpin-derived peptide substrates for investigating the substrate specificity of human tissue kallikreins hK1 and hK2. J. Biol. Chem. 272, 29590–29595 (1997).
Rehault, S. et al. Design of new and sensitive fluorogenic substrates for human kallikrein hK3 (prostate-specific antigen) derived from semenogelin sequences. Biochim. Biophys. Acta 1596, 55–62 (2002).
Yang, C. F. et al. Design of synthetic hexapeptide substrates for prostate-specific antigen using single-position minilibraries. J. Pept. Res. 54, 444–448 (1999).
Rajapakse, S., Ogiwara, K., Takano, N., Moriyama, A. & Takahashi, T. Biochemical characterization of human kallikrein 8 and its possible involvement in the degradation of extracellular matrix proteins. FEBS Lett. 579, 6879–6884 (2005).
Rajapakse, S. & Takahashi, T. Expression and enzymatic characterization of recombinant human kallikrein 14. Zoolog Sci. 24, 774–780 (2007).
Memari, N., Jiang, W., Diamandis, E. P. & Luo, L. Y. Enzymatic properties of human kallikrein-related peptidase 12 (KLK12). Biol. Chem. 388, 427–435 (2007).
Kapadia, C., Ghosh, M. C., Grass, L. & Diamandis, E. P. Human kallikrein 13 involvement in extracellular matrix degradation. Biochem. Biophys. Res. Commun. 323, 1084–1090 (2004).
Magklara, A. et al. Characterization of the enzymatic activity of human kallikrein 6: autoactivation, substrate specificity, and regulation by inhibitors. Biochem. Biophys. Res. Commun. 307, 948–955 (2003).
Debela, M. et al. Structures and specificity of the human kallikrein-related peptidases KLK 4, 5, 6, and 7. Biol. Chem. 389, 623–632 (2008). This is the first report on the characterization of the crystal structure of four KLKs.
Debela, M. et al. Structural basis of the zinc inhibition of human tissue kallikrein 5. J. Mol. Biol. 373, 1017–1031 (2007).
Debela, M. et al. Chymotryptic specificity determinants in the 1.0Å structure of the zinc-inhibited human tissue kallikrein 7. Proc. Natl Acad. Sci. USA 104, 16086–16091 (2007).
Debela, M. et al. Crystal structures of human tissue kallikrein 4: activity modulation by a specific zinc binding site. J. Mol. Biol. 362, 1094–1107 (2006).
Katz, B. A., Liu, B., Barnes, M. & Springman, E. B. Crystal structure of recombinant human tissue kallikrein at 2.0Å resolution. Protein Sci. 7, 875–885 (1998).
Kishi, T. et al. Crystal structure of neuropsin, a hippocampal protease involved in kindling epileptogenesis. J. Biol. Chem. 274, 4220–4224 (1999).
Bernett, M. J. et al. Crystal structure and biochemical characterization of human kallikrein 6 reveals that a trypsin-like kallikrein is expressed in the central nervous system. J. Biol. Chem. 277, 24562–24570 (2002).
Laxmikanthan, G. et al. 1.70Å X-ray structure of human apo kallikrein 1: structural changes upon peptide inhibitor/substrate binding. Proteins 58, 802–814 (2005).
Polgar, L. The catalytic triad of serine peptidases. Cell. Mol. Life Sci. 62, 2161–2172 (2005).
Shaw, J. L. & Diamandis, E. P. Distribution of 15 human kallikreins in tissues and biological fluids. Clin. Chem. 53, 1423–1432 (2007).
Borgono, C. A. et al. A potential role for multiple tissue kallikrein serine proteases in epidermal desquamation. J. Biol. Chem. 282, 3640–3652 (2007).
Komatsu, N. et al. Quantification of human tissue kallikreins in the stratum corneum: dependence on age and gender. J. Invest. Dermatol. 125, 1182–1189 (2005).
Komatsu, N. et al. Multiple tissue kallikrein mRNA and protein expression in normal skin and skin diseases. Br. J. Dermatol. 153, 274–281 (2005).
Komatsu, N. et al. Quantification of eight tissue kallikreins in the stratum corneum and sweat. J. Invest. Dermatol. 126, 925–929 (2006).
Brattsand, M. & Egelrud, T. Purification, molecular cloning, and expression of a human stratum corneum trypsin-like serine protease with possible function in desquamation. J. Biol. Chem. 274, 30033–30040 (1999).
Hansson, L. et al. Cloning, expression, and characterization of stratum corneum chymotryptic enzyme. A skin-specific human serine proteinase. J. Biol. Chem. 269, 19420–19426 (1994).
Brattsand, M., Stefansson, K., Lundh, C., Haasum, Y. & Egelrud, T. A proteolytic cascade of kallikreins in the stratum corneum. J. Invest. Dermatol. 124, 198–203 (2005). The paper elucidates the proteolytic interplay among active human KLK5, KLK7 and KLK14 in the epidermis.
Eissa, A., Amodeo, V., Smith, C. R. & Diamandis, E. P. Kallikrein-related peptidase-8 (KLK8) is an active serine protease in human epidermis and sweat and is involved in a skin barrier proteolytic cascade. J. Biol. Chem. 286, 687–706 (2011).
Stefansson, K., Brattsand, M., Ny, A., Glas, B. & Egelrud, T. Kallikrein-related peptidase 14 may be a major contributor to trypsin-like proteolytic activity in human stratum corneum. Biol. Chem. 387, 761–768 (2006).
Eissa, A. & Diamandis, E. P. Human tissue kallikreins as promiscuous modulators of homeostatic skin barrier functions. Biol. Chem. 389, 669–680 (2008).
Kirihara, T. et al. Prolonged recovery of ultraviolet B-irradiated skin in neuropsin (KLK8)-deficient mice. Br. J. Dermatol. 149, 700–706 (2003).
Hachem, J. P. et al. Sustained serine proteases activity by prolonged increase in pH leads to degradation of lipid processing enzymes and profound alterations of barrier function and stratum corneum integrity. J. Invest. Dermatol. 125, 510–520 (2005).
Hachem, J. P. et al. Serine protease activity and residual LEKTI expression determine phenotype in Netherton syndrome. J. Invest. Dermatol. 126, 1609–1621 (2006).
Yamasaki, K. et al. Increased serine protease activity and cathelicidin promotes skin inflammation in rosacea. Nature Med. 13, 975–980 (2007). This paper reports the discovery of the active roles of KLK5 and KLK7 in the pathogenesis of rosacea.
Kanada, K. N., Nakatsuji, T. & Gallo, R. L. Doxycycline indirectly inhibits proteolytic activation of tryptic kallikrein-related peptidases and activation of cathelicidin. J. Invest. Dermatol. 132, 1435–1442 (2012).
Morizane, S., Yamasaki, K., Kabigting, F. D. & Gallo, R. L. Kallikrein expression and cathelicidin processing are independently controlled in keratinocytes by calcium, vitamin D3, and retinoic acid. J. Invest. Dermatol. 130, 1297–1306 (2010).
Ekholm, E. & Egelrud, T. Stratum corneum chymotryptic enzyme in psoriasis. Arch. Dermatol. Res. 291, 195–200 (1999).
Descargues, P. et al. Spink5-deficient mice mimic Netherton syndrome through degradation of desmoglein 1 by epidermal protease hyperactivity. Nature Genet. 37, 56–65 (2005). This paper reports the identification of LEKTI as a key regulator of epidermal protease activity, and the degradation of DSG1 as the primary pathogenic event in Netherton syndrome.
Bin, L., Kim, B. E., Hall, C. F., Leach, S. M. & Leung, D. Y. Inhibition of transcription factor specificity protein 1 alters the gene expression profile of keratinocytes leading to upregulation of kallikrein-related peptidases and thymic stromal lymphopoietin. J. Invest. Dermatol. 131, 2213–2222 (2011).
Lu, J. et al. Transcriptional profiling of keratinocytes reveals a vitamin D-regulated epidermal differentiation network. J. Invest. Dermatol. 124, 778–785 (2005).
Hovnanian, A. Netherton syndrome: skin inflammation and allergy by loss of protease inhibition. Cell Tissue Res. 351, 289–300 (2013).
Chavanas, S. et al. Mutations in SPINK5, encoding a serine protease inhibitor, cause Netherton syndrome. Nature Genet. 25, 141–142 (2000).
Komatsu, N. et al. Elevated stratum corneum hydrolytic activity in Netherton syndrome suggests an inhibitory regulation of desquamation by SPINK5-derived peptides. J. Invest. Dermatol. 118, 436–443 (2002).
Bitoun, E. et al. LEKTI proteolytic processing in human primary keratinocytes, tissue distribution and defective expression in Netherton syndrome. Hum. Mol. Genet. 12, 2417–2430 (2003).
Deraison, C. et al. LEKTI fragments specifically inhibit KLK5, KLK7, and KLK14 and control desquamation through a pH-dependent interaction. Mol. Biol. Cell 18, 3607–3619 (2007).
Fortugno, P. et al. The 420K LEKTI variant alters LEKTI proteolytic activation and results in protease deregulation: implications for atopic dermatitis. Hum. Mol. Genet. 21, 4187–4200 (2012).
Furio, L. & Hovnanian, A. When activity requires breaking up: LEKTI proteolytic activation cascade for specific proteinase inhibition. J. Invest. Dermatol. 131, 2169–2173 (2011).
Ishida-Yamamoto, A. et al. LEKTI is localized in lamellar granules, separated from KLK5 and KLK7, and is secreted in the extracellular spaces of the superficial stratum granulosum. J. Invest. Dermatol. 124, 360–366 (2005).
Wang, S. et al. SPINK5 knockdown in organotypic human skin culture as a model system for Netherton syndrome: effect of genetic inhibition of serine proteases kallikrein 5 and kallikrein 7. Exp. Dermatol. 23, 524–526 (2014).
Sales, K. U. et al. Matriptase initiates activation of epidermal pro-kallikrein and disease onset in a mouse model of Netherton syndrome. Nature Genet. 42, 676–683 (2010). The study implicated a crucial pathogenic matriptase–pro-KLK pathway in several inflammatory skin diseases.
Briot, A. et al. Kallikrein 5 induces atopic dermatitis-like lesions through PAR2-mediated thymic stromal lymphopoietin expression in Netherton syndrome. J. Exp. Med. 206, 1135–1147 (2009).
Briot, A. et al. Par2 inactivation inhibits early production of TSLP, but not cutaneous inflammation, in Netherton syndrome adult mouse model. J. Invest. Dermatol. 130, 2736–2742 (2010). This study supports the role of the KLK5–PAR2 cascade in TSLP-mediated early proallergic signalling, by using a Spink5−/− and Par2−/− double knockout mouse model.
Furio, L. et al. Transgenic kallikrein 5 mice reproduce major cutaneous and systemic hallmarks of Netherton syndrome. J. Exp. Med. 211, 499–513 (2014). The paper reports the generation of a new transgenic murine model expressing human KLK5 in the granular layer of the epidermis, and validates KLK5 as a major therapeutic target for Netherton Syndrome.
Komatsu, N. et al. Human tissue kallikrein expression in the stratum corneum and serum of atopic dermatitis patients. Exp. Dermatol. 16, 513–519 (2007).
Tanaka, R. J., Ono, M. & Harrington, H. A. Skin barrier homeostasis in atopic dermatitis: feedback regulation of kallikrein activity. PLoS ONE 6, e19895 (2011).
Vasilopoulos, Y. et al. The 3′-UTR AACCins5874 in the stratum corneum chymotryptic enzyme gene (SCCE/KLK7), associated with atopic dermatitis; causes an increased mRNA expression without altering its stability. J. Dermatol. Sci. 61, 131–133 (2011).
Voegeli, R. et al. Increased mass levels of certain serine proteases in the stratum corneum in acute eczematous atopic skin. Int. J. Cosmet. Sci. 33, 560–565 (2011).
Ny, A. & Egelrud, T. Epidermal hyperproliferation and decreased skin barrier function in mice overexpressing stratum corneum chymotryptic enzyme. Acta Derm. Venereol. 84, 18–22 (2004).
Stefansson, K. et al. Activation of proteinase-activated receptor-2 by human kallikrein-related peptidases. J. Invest. Dermatol. 128, 18–25 (2008).
Gottlieb, A. B. Psoriasis: emerging therapeutic strategies. Nature Rev. Drug Discov. 4, 19–34 (2005).
Nestle, F. O., Kaplan, D. H. & Barker, J. Psoriasis. N. Engl. J. Med. 361, 496–509 (2009).
Eissa, A. et al. Serum kallikrein-8 correlates with skin activity, but not psoriatic arthritis, in patients with psoriatic disease. Clin. Chem. Lab. Med. 51, 317–325 (2013).
Komatsu, N. et al. Aberrant human tissue kallikrein levels in the stratum corneum and serum of patients with psoriasis: dependence on phenotype, severity and therapy. Br. J. Dermatol. 156, 875–883 (2007).
Ainali, C. et al. Transcriptome classification reveals molecular subtypes in psoriasis. BMC Genomics 13, 472 (2012).
Kishibe, M. et al. Kallikrein-related peptidase 8-dependent skin wound healing is associated with upregulation of kallikrein-related peptidase 6 and PAR2. J. Invest. Dermatol. 132, 1717–1724 (2012). This is a critical study on the roles of KLK8 in wound healing.
Kishibe, M. et al. Kallikrein 8 is involved in skin desquamation in cooperation with other kallikreins. J. Biol. Chem. 282, 5834–5841 (2007).
Fischer, J. et al. Characterization of Spink6 in mouse skin: the conserved inhibitor of kallikrein-related peptidases is reduced by barrier injury. J. Invest. Dermatol. 134, 1305–1312 (2013).
Wang, X. et al. AP-2α: a regulator of EGF receptor signaling and proliferation in skin epidermis. J. Cell Biol. 172, 409–421 (2006).
Shingaki, K. et al. Molecular mechanism of kallikrein-related peptidase 8/neuropsin-induced hyperkeratosis in inflamed skin. Br. J. Dermatol. 163, 466–475 (2010).
Nylander-Lundqvist, E. & Egelrud, T. Formation of active IL-1β from pro-IL-1β catalyzed by stratum corneum chymotryptic enzyme in vitro. Acta Derm. Venereol. 77, 203–206 (1997).
Culp, B. & Scheinfeld, N. Rosacea: a review. P. T. 34, 38–45 (2009).
Coda, A. B. et al. Cathelicidin, kallikrein 5, and serine protease activity is inhibited during treatment of rosacea with azelaic acid 15% gel. J. Am. Acad. Dermatol. 69, 570–577 (2013).
Glukhen'kii, B. T. & Snitsarenko, O. V. The kallikrein-kinin system in rosacea patients. Vestn. Dermatol. Venerol. 6, 30–32 (in Russian) (1985).
Meyer-Hoffert, U. & Schroder, J. M. Epidermal proteases in the pathogenesis of rosacea. J. Investig. Dermatol. Symp. Proc. 15, 16–23 (2011).
Yamasaki, K. & Gallo, R. L. Rosacea as a disease of cathelicidins and skin innate immunity. J. Investig. Dermatol. Symp. Proc. 15, 12–15 (2011).
Yamasaki, K. et al. TLR2 expression is increased in rosacea and stimulates enhanced serine protease production by keratinocytes. J. Invest. Dermatol. 131, 688–697 (2011).
Kasetty, G. et al. The C-terminal sequence of several human serine proteases encodes host defense functions. J. Innate Immun. 3, 471–482 (2011).
Emami, N., Deperthes, D., Malm, J. & Diamandis, E. P. Major role of human KLK14 in seminal clot liquefaction. J. Biol. Chem. 283, 19561–19569 (2008).
Emami, N. & Diamandis, E. P. Potential role of multiple members of the kallikrein-related peptidase family of serine proteases in activating latent TGFβ1 in semen. Biol. Chem. 391, 85–95 (2010).
Emami, N. & Diamandis, E. P. Human kallikrein-related peptidase 14 (KLK14) is a new activator component of the KLK proteolytic cascade. Possible function in seminal plasma and skin. J. Biol. Chem. 283, 3031–3041 (2008).
Veveris-Lowe, T. L., Kruger, S. J., Walsh, T., Gardiner, R. A. & Clements, J. A. Seminal fluid characterization for male fertility and prostate cancer: kallikrein-related serine proteases and whole proteome approaches. Semin. Thromb. Hemost. 33, 87–99 (2007).
Michael, I. P. et al. Human tissue kallikrein 5 is a member of a proteolytic cascade pathway involved in seminal clot liquefaction and potentially in prostate cancer progression. J. Biol. Chem. 281, 12743–12750 (2006).
Costello, L. C., Feng, P., Milon, B., Tan, M. & Franklin, R. B. Role of zinc in the pathogenesis and treatment of prostate cancer: critical issues to resolve. Prostate Cancer Prostat. Dis. 7, 111–117 (2004).
Mattsson, J. M. et al. Proteolytic activity of prostate-specific antigen (PSA) towards protein substrates and effect of peptides stimulating PSA activity. PLoS ONE 9, e107819 (2014).
Logothetis, C. J. & Lin, S. H. Osteoblasts in prostate cancer metastasis to bone. Nature Rev. Cancer 5, 21–28 (2005).
Avgeris, M., Stravodimos, K. & Scorilas, A. Kallikrein-related peptidase 4 gene (KLK4) in prostate tumors: quantitative expression analysis and evaluation of its clinical significance. Prostate 71, 1780–1789 (2011).
Seiz, L. et al. Polyclonal antibodies against kallikrein-related peptidase 4 (KLK4): immunohistochemical assessment of KLK4 expression in healthy tissues and prostate cancer. Biol. Chem. 391, 391–401 (2010).
Veveris-Lowe, T. L. et al. Kallikrein 4 (hK4) and prostate-specific antigen (PSA) are associated with the loss of E-cadherin and an epithelial-mesenchymal transition (EMT)-like effect in prostate cancer cells. Endocr. Relat. Cancer 12, 631–643 (2005).
Ramsay, A. J. et al. Kallikrein-related peptidase 4 (KLK4) initiates intracellular signaling via protease-activated receptors (PARs). KLK4 and PAR-2 are co-expressed during prostate cancer progression. J. Biol. Chem. 283, 12293–12304 (2008).
Gratio, V. et al. Kallikrein-related peptidase 4: a new activator of the aberrantly expressed protease-activated receptor 1 in colon cancer cells. Am. J. Pathol. 176, 1452–1461 (2010).
Mize, G. J., Wang, W. & Takayama, T. K. Prostate-specific kallikreins-2 and -4 enhance the proliferation of DU-145 prostate cancer cells through protease-activated receptors-1 and -2. Mol. Cancer Res. 6, 1043–1051 (2008).
Wang, W., Mize, G. J., Zhang, X. & Takayama, T. K. Kallikrein-related peptidase-4 initiates tumor-stroma interactions in prostate cancer through protease-activated receptor-1. Int. J. Cancer 126, 599–610 (2010).
Gao, J. et al. Kallikrein 4 is a potential mediator of cellular interactions between cancer cells and osteoblasts in metastatic prostate cancer. Prostate 67, 348–360 (2007).
Jin, Y. et al. Molecular circuit involving KLK4 integrates androgen and mTOR signaling in prostate cancer. Proc. Natl Acad. Sci. USA 110, E2572–E2581 (2013). This study validates KLK4 as a promising therapeutic target for prostate cancer.
Lose, F. et al. Genetic association of the KLK4 locus with risk of prostate cancer. PLoS ONE 7, e44520 (2012).
Madeddu, P., Emanueli, C. & El-Dahr, S. Mechanisms of disease: the tissue kallikrein-kinin system in hypertension and vascular remodeling. Nature Clin. Pract. Nephrol. 3, 208–221 (2007).
Davis, W. J. & Feldman, B. R. Treatment of asthma with theophylline and β adrenergic agents. Cutis 17, 1091–1098 (1976).
O'Riordan, T. G., Weinstein, M. D., Abraham, W. M. & Forteza, R. Elevated tissue kallikrein activity in airway secretions from patients with tracheobronchitis associated with prolonged mechanical ventilation. Lung 181, 237–244 (2003).
Lauredo, I. T., Forteza, R. M., Botvinnikova, Y. & Abraham, W. M. Leukocytic cell sources of airway tissue kallikrein. Am. J. Physiol. Lung Cell. Mol. Physiol. 286, L734–L740 (2004).
Forteza, R., Lauredo, I., Abraham, W. M. & Conner, G. E. Bronchial tissue kallikrein activity is regulated by hyaluronic acid binding. Am. J. Respir. Cell. Mol. Biol. 21, 666–674 (1999).
Forteza, R. et al. Hyaluronan serves a novel role in airway mucosal host defense. FASEB J. 15, 2179–2186 (2001).
Casalino-Matsuda, S. M., Monzon, M. E., Conner, G. E., Salathe, M. & Forteza, R. M. Role of hyaluronan and reactive oxygen species in tissue kallikrein-mediated epidermal growth factor receptor activation in human airways. J. Biol. Chem. 279, 21606–21616 (2004). An in-depth mechanistic study on the signalling properties of KLK1 in human airways.
Casalino-Matsuda, S. M., Monzon, M. E. & Forteza, R. M. Epidermal growth factor receptor activation by epidermal growth factor mediates oxidant-induced goblet cell metaplasia in human airway epithelium. Am. J. Respir. Cell. Mol. Biol. 34, 581–591 (2006).
Christiansen, S. C. et al. Experimental rhinovirus infection increases human tissue kallikrein activation in allergic subjects. Int. Arch. Allergy Immunol. 147, 299–304 (2008).
Sexton, D. J. et al. Specific inhibition of tissue kallikrein 1 with a human monoclonal antibody reveals a potential role in airway diseases. Biochem. J. 422, 383–392 (2009).
Gondzik, V., Weber, W. M. & Awayda, M. S. Coupling of epithelial Na+ and Cl− channels by direct and indirect activation by serine proteases. Am. J. Physiol. Cell Physiol. 303, C936–C946 (2012).
Patel, A. B., Chao, J. & Palmer, L. G. Tissue kallikrein activation of the epithelial Na channel. Am. J. Physiol. Renal Physiol. 303, F540–F550 (2012).
Topala, C. N., Bindels, R. J. & Hoenderop, J. G. Regulation of the epithelial calcium channel TRPV5 by extracellular factors. Curr. Opin. Nephrol. Hypertens. 16, 319–324 (2007).
El Moghrabi, S. et al. Tissue kallikrein permits early renal adaptation to potassium load. Proc. Natl Acad. Sci. USA 107, 13526–13531 (2010).
Picard, N. et al. Defective ENaC processing and function in tissue kallikrein-deficient mice. J. Biol. Chem. 283, 4602–4611 (2008).
Li, Q. D., Li, F. J., Liu, X. C. & Jiang, H. KLK1 A1789G gene polymorphism and the risk of coronary artery stenosis in the Chinese population. Genet. Mol. Res. 12, 1636–1645 (2013).
Azizi, M. et al. Arterial and renal consequences of partial genetic deficiency in tissue kallikrein activity in humans. J. Clin. Invest. 115, 780–787 (2005).
Carretero, O. A. Vascular remodeling and the kallikrein-kinin system. J. Clin. Invest. 115, 588–591 (2005).
Ponticelli, C. & Meroni, P. L. Kallikreins and lupus nephritis. J. Clin. Invest. 119, 768–771 (2009).
Li, Q. Z. et al. The lupus-susceptibility gene kallikrein downmodulates antibody-mediated glomerulonephritis. Genes Immun. 10, 503–508 (2009).
Simmer, J. P. et al. Purification, characterization, and cloning of enamel matrix serine proteinase 1. J. Dent. Res. 77, 377–386 (1998).
Bartlett, J. D. Dental enamel development: proteinases and their enamel matrix substrates. ISRN Dent. 2013, 684607 (2013).
Cho, A. et al. TGF-β regulates enamel mineralization and maturation through KLK4 expression. PLoS ONE 8, e82267 (2013).
Smith, C. E. et al. Relationships between protein and mineral during enamel development in normal and genetically altered mice. Eur. J. Oral Sci. 119 (Suppl. 1), 125–135 (2011).
Hart, P. S. et al. Mutation in kallikrein 4 causes autosomal recessive hypomaturation amelogenesis imperfecta. J. Med. Genet. 41, 545–549 (2004).
Wang, S. K. et al. Novel KLK4 and MMP20 mutations discovered by whole-exome sequencing. J. Dent. Res. 92, 266–271 (2013).
Bando, Y. et al. Implications of protease M/neurosin in myelination during experimental demyelination and remyelination. Neurosci. Lett. 405, 175–180 (2006).
Yoshida, S. Klk8, a multifunctional protease in the brain and skin: analysis of knockout mice. Biol. Chem. 391, 375–380 (2010).
Terayama, R. et al. Neuropsin promotes oligodendrocyte death, demyelination and axonal degeneration after spinal cord injury. Neuroscience 148, 175–187 (2007).
Terayama, R., Bando, Y., Takahashi, T. & Yoshida, S. Differential expression of neuropsin and protease M/neurosin in oligodendrocytes after injury to the spinal cord. Glia 48, 91–101 (2004).
Nakamura, Y., Tamura, H., Horinouchi, K. & Shiosaka, S. Role of neuropsin in formation and maturation of Schaffer-collateral L1cam-immunoreactive synaptic boutons. J. Cell Sci. 119, 1341–1349 (2006).
Tamura, H. et al. Neuropsin is essential for early processes of memory acquisition and Schaffer collateral long-term potentiation in adult mouse hippocampus in vivo. J. Physiol. 570, 541–551 (2006).
Komai, S. et al. Neuropsin regulates an early phase of schaffer-collateral long-term potentiation in the murine hippocampus. Eur. J. Neurosci. 12, 1479–1486 (2000).
Matsumoto-Miyai, K. et al. NMDA-dependent proteolysis of presynaptic adhesion molecule L1 in the hippocampus by neuropsin. J. Neurosci. 23, 7727–7736 (2003).
Attwood, B. K. et al. Neuropsin cleaves EphB2 in the amygdala to control anxiety. Nature 473, 372–375 (2011). This paper reports a novel neuronal pathway linking stress-induced KLK8-driven proteolysis of EPHB2 in the amygdala to anxiety.
Tamura, H., Kawata, M., Hamaguchi, S., Ishikawa, Y. & Shiosaka, S. Processing of neuregulin-1 by neuropsin regulates GABAergic neuron to control neural plasticity of the mouse hippocampus. J. Neurosci. 32, 12657–12672 (2012). This paper presents critical insights into the roles of KLK8 in human cognition and mental disorders, such as schizophrenia and bipolar disorder.
Mei, L. & Xiong, W. C. Neuregulin 1 in neural development, synaptic plasticity and schizophrenia. Nature Rev. Neurosci. 9, 437–452 (2008).
Izumi, A. et al. Genetic variations of human neuropsin gene and psychiatric disorders: polymorphism screening and possible association with bipolar disorder and cognitive functions. Neuropsychopharmacology 33, 3237–3245 (2008).
Murakami, K. et al. In vivo analysis of kallikrein-related peptidase 6 (KLK6) function in oligodendrocyte development and the expression of myelin proteins. Neuroscience 236, 1–11 (2013).
Yousef, G. M., Kishi, T. & Diamandis, E. P. Role of kallikrein enzymes in the central nervous system. Clin. Chim. Acta 329, 1–8 (2003).
Aoyagi, T. et al. Deficiency of kallikrein-like enzyme activities in cerebral tissue of patients with Alzheimer's disease. Experientia 46, 94–97 (1990).
Ogawa, K. et al. Localization of a novel type trypsin-like serine protease, neurosin, in brain tissues of Alzheimer's disease and Parkinson's disease. Psychiatry Clin. Neurosci. 54, 419–426 (2000).
Zarghooni, M. et al. Decreased concentration of human kallikrein 6 in brain extracts of Alzheimer's disease patients. Clin. Biochem. 35, 225–231 (2002).
Shulman, J. M. & De Jager, P. L. Evidence for a common pathway linking neurodegenerative diseases. Nature Genet. 41, 1261–1262 (2009).
Iwata, A. et al. α-synuclein degradation by serine protease neurosin: implication for pathogenesis of synucleinopathies. Hum. Mol. Genet. 12, 2625–2635 (2003).
Kasai, T. et al. Cleavage of normal and pathological forms of α-synuclein by neurosin in vitro. Neurosci. Lett. 436, 52–56 (2008).
Tatebe, H. et al. Extracellular neurosin degrades α-synuclein in cultured cells. Neurosci. Res. 67, 341–346 (2010).
Spencer, B. et al. Lentivirus mediated delivery of neurosin promotes clearance of wild-type α-synuclein and reduces the pathology in an α-synuclein model of LBD. Mol. Ther. 21, 31–41 (2013).
Blaber, S. I. et al. Targeting kallikrein 6 proteolysis attenuates CNS inflammatory disease. FASEB J. 18, 920–922 (2004).
Scarisbrick, I. A. et al. Kallikrein 6 regulates early CNS demyelination in a viral model of multiple sclerosis. Brain Pathol. 22, 709–722 (2012).
Blaber, S. I. et al. Enzymatic properties of rat myelencephalon-specific protease. Biochemistry 41, 1165–1173 (2002).
Terayama, R., Bando, Y., Yamada, M. & Yoshida, S. Involvement of neuropsin in the pathogenesis of experimental autoimmune encephalomyelitis. Glia 52, 108–118 (2005).
Scarisbrick, I. A. et al. Kallikrein 6 is a novel molecular trigger of reactive astrogliosis. Biol. Chem. 393, 355–367 (2012).
Noorbakhsh, F. et al. Proteinase-activated receptor 2 modulates neuroinflammation in experimental autoimmune encephalomyelitis and multiple sclerosis. J. Exp. Med. 203, 425–435 (2006).
Koistinen, H. et al. Novel small molecule inhibitors for prostate-specific antigen. Prostate 68, 1143–1151 (2008).
Tan, X. et al. 1,2,4-triazole derivatives as transient inactivators of kallikreins involved in skin diseases. Bioorg. Med. Chem. Lett. 23, 4547–4551 (2013).
Tan, X. et al. Identification by in silico and in vitro screenings of small organic molecules acting as reversible inhibitors of kallikreins. Eur. J. Med. Chem. 70, 661–668 (2013).
Liang, G. et al. Human kallikrein 6 inhibitors with a para-amidobenzylanmine P1 group identified through virtual screening. Bioorg. Med. Chem. Lett. 22, 2450–2455 (2012).
Flohr, S. et al. Kallikrein 7 modulators WO 2009000878 A1. (2008).
Freitas, R. F. et al. Isomannide derivatives as new class of inhibitors for human kallikrein 7. Bioorg. Med. Chem. Lett. 22, 6072–6075 (2012).
Singh, P., LeBeau, A. M., Lilja, H., Denmeade, S. R. & Isaacs, J. T. Molecular insights into substrate specificity of prostate specific antigen through structural modeling. Proteins 77, 984–993 (2009).
Vlieghe, P., Lisowski, V., Martinez, J. & Khrestchatisky, M. Synthetic therapeutic peptides: science and market. Drug Discov. Today 15, 40–56 (2010).
LeBeau, A. M., Banerjee, S. R., Pomper, M. G., Mease, R. C. & Denmeade, S. R. Optimization of peptide-based inhibitors of prostate-specific antigen (PSA) as targeted imaging agents for prostate cancer. Bioorg. Med. Chem. 17, 4888–4893 (2009).
LeBeau, A. M., Singh, P., Isaacs, J. T. & Denmeade, S. R. Potent and selective peptidyl boronic acid inhibitors of the serine protease prostate-specific antigen. Chem. Biol. 15, 665–674 (2008).
Evans, D. M. et al. Synthetic inhibitors of human tissue kallikrein. Immunopharmacology 32, 117–118 (1996).
Koistinen, H., Hekim, C., Wu, P., Narvanen, A. & Stenman, U. H. Evaluation of peptides as protease inhibitors and stimulators. Methods Mol. Biol. 1088, 147–158 (2014).
Pakkala, M. et al. Activity and stability of human kallikrein-2-specific linear and cyclic peptide inhibitors. J. Pept. Sci. 13, 348–353 (2007).
Huang, X. et al. Modulation of recombinant human prostate-specific antigen: activation by Hofmeister salts and inhibition by azapeptides. Appendix: thermodynamic interpretation of the activation by concentrated salts. Biochemistry 40, 11734–11741 (2001).
Evans, D.M., Allans, C.E., Horton, J. & Rooker, D.P. Aminopyridine derivatives WO 2009133348 A1. (2009).
Vasileiou, Z. et al. Synthesis of the proteinase inhibitor LEKTI domain 6 by the fragment condensation method and regioselective disulfide bond formation. Biopolymers 94, 339–349 (2010).
de Veer, S. J. et al. Mechanism-based selection of a potent kallikrein-related peptidase 7 inhibitor from a versatile library based on the sunflower trypsin inhibitor SFTI-1. Biopolymers (2013).
Swedberg, J. E. et al. Mastering the canonical loop of serine protease inhibitors: enhancing potency by optimising the internal hydrogen bond network. PLoS ONE 6, e19302 (2011).
Felber, L. M. et al. Mutant recombinant serpins as highly specific inhibitors of human kallikrein 14. FEBS J. 273, 2505–2514 (2006).
Cloutier, S. M. et al. Development of recombinant inhibitors specific to human kallikrein 2 using phage-display selected substrates. Eur. J. Biochem. 271, 607–613 (2004).
Cha, E. & Fong, L. Immunotherapy for prostate cancer: biology and therapeutic approaches. J. Clin. Oncol. 29, 3677–3685 (2011).
Madan, R. A. & Gulley, J. L. The current and emerging role of immunotherapy in prostate cancer. Clin. Genitourin. Cancer 8, 10–16 (2010).
Madan, R. A. & Gulley, J. L. Therapeutic cancer vaccine fulfills the promise of immunotherapy in prostate cancer. Immunotherapy 3, 27–31 (2011).
Di Lorenzo, G., Buonerba, C. & Kantoff, P. W. Immunotherapy for the treatment of prostate cancer. Nature Rev. Clin. Oncol. 8, 551–561 (2011).
Madan, R. A., Arlen, P. M., Mohebtash, M., Hodge, J. W. & Gulley, J. L. Prostvac-VF: a vector-based vaccine targeting PSA in prostate cancer. Expert Opin. Investig. Drugs 18, 1001–1011 (2009).
Kantoff, P. W. et al. Overall survival analysis of a phase II randomized controlled trial of a Poxviral-based PSA-targeted immunotherapy in metastatic castration-resistant prostate cancer. J. Clin. Oncol. 28, 1099–1105 (2010).
Madan, R. A. & Gulley, J. L. Sipuleucel-T: harbinger of a new age of therapeutics for prostate cancer. Expert Rev. Vaccines 10, 141–150 (2011).
Sonpavde, G. et al. The role of sipuleucel-T in therapy for castration-resistant prostate cancer: a critical analysis of the literature. Eur. Urol. 61, 639–647 (2012).
Arlen, P. M., Mohebtash, M., Madan, R. A. & Gulley, J. L. Promising novel immunotherapies and combinations for prostate cancer. Future Oncol. 5, 187–196 (2009).
Bitting, R. L. & Armstrong, A. J. Potential predictive biomarkers for individualizing treatment for men with castration-resistant prostate cancer. Cancer J. 19, 25–33 (2013).
May, K. F. et al. Prostate cancer immunotherapy. Clin. Cancer Res. 17, 5233–5238 (2011).
Schweizer, M. T. & Drake, C. G. Immunotherapy for prostate cancer: recent developments and future challenges. Cancer Metastasis Rev. 33, 641–655 (2014).
Thakur, A., Vaishampayan, U. & Lum, L. G. Immunotherapy and immune evasion in prostate cancer. Cancers (Basel) 5, 569–590 (2013).
auf dem Keller, U., Doucet, A. & Overall, C. M. Protease research in the era of systems biology. Biol. Chem. 388, 1159–1162 (2007).
Beaufort, N. et al. Interdependence of kallikrein-related peptidases in proteolytic networks. Biol. Chem. 391, 581–587 (2010).
Ohler, A., Debela, M., Wagner, S., Magdolen, V. & Becker-Pauly, C. Analyzing the protease web in skin: meprin metalloproteases are activated specifically by KLK4, 5 and 8 vice versa leading to processing of proKLK7 thereby triggering its activation. Biol. Chem. 391, 455–460 (2010).
Nomura, D. K., Dix, M. M. & Cravatt, B. F. Activity-based protein profiling for biochemical pathway discovery in cancer. Nature Rev. Cancer 10, 630–638 (2010).
Jeffery, D. A. & Bogyo, M. Chemical proteomics and its application to drug discovery. Drug Discov. Today 9, S19–S26 (2004).
Schilling, O., Huesgen, P. F., Barre, O., Auf dem Keller, U. & Overall, C. M. Characterization of the prime and non-prime active site specificities of proteases by proteome-derived peptide libraries and tandem mass spectrometry. Nature Protoc. 6, 111–120 (2011).
Kleifeld, O. et al. Isotopic labeling of terminal amines in complex samples identifies protein N-termini and protease cleavage products. Nature Biotech. 28, 281–288 (2010).
Doucet, A. & Overall, C. M. Protease proteomics: revealing protease in vivo functions using systems biology approaches. Mol. Aspects Med. 29, 339–358 (2008).
Turk, B. Targeting proteases: successes, failures and future prospects. Nature Rev. Drug Discov. 5, 785–799 (2006).
Walker, B. & Lynas, J. F. Strategies for the inhibition of serine proteases. Cell. Mol. Life Sci. 58, 596–624 (2001).
Pinto, D. J. et al. Discovery of 1-(4-methoxyphenyl)-7-oxo-6-(4-(2-oxopiperidin-1-yl)phenyl)-4,5,6,7- tetrahydro-1H-pyrazolo[3,4-c]pyridine-3-carboxamide (apixaban, BMS-562247), a highly potent, selective, efficacious, and orally bioavailable inhibitor of blood coagulation factor Xa. J. Med. Chem. 50, 5339–5356 (2007).
Pinto, D. J., Qiao, J. X. & Knabb, R. M. The emergence of factor Xa inhibitors for the treatment of cardiovascular diseases: a patent review. Expert Opin. Ther. Pat. 22, 645–661 (2012).
Latham, P. W. Therapeutic peptides revisited. Nature Biotech. 17, 755–757 (1999).
Overall, C. M. & Lopez-Otin, C. Strategies for MMP inhibition in cancer: innovations for the post-trial era. Nature Rev. Cancer 2, 657–672 (2002).
Meneton, P. et al. Cardiovascular abnormalities with normal blood pressure in tissue kallikrein-deficient mice. Proc. Natl Acad. Sci. USA 98, 2634–2639 (2001).
Pons, S. et al. Tissue kallikrein deficiency aggravates cardiac remodelling and decreases survival after myocardial infarction in mice. Eur. J. Heart Fail. 10, 343–351 (2008).
Simmer, J. P., Hu, Y., Lertlam, R., Yamakoshi, Y. & Hu, J. C. Hypomaturation enamel defects in Klk4 knockout/LacZ knockin mice. J. Biol. Chem. 284, 19110–19121 (2009).
Acknowledgements
G.P. would like to acknowledge the support of the Ontario Institute for Cancer Research and its funding from the Government of Ontario, Canada.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Supplementary information
Supplementary information S1 (table)
Summary of the % nucleotide sequence identities between KLK-related peptidases* (PDF 120 kb)
Supplementary information S2 (table)
Diagrammatic summary of the preferred substrate specificities of the human KLK peptidases according to the MEROPS Peptidase Database* (PDF 111 kb)
Supplementary information S3 (figure)
Structure of other major serine proteases in standard orientation. (PDF 375 kb)
Glossary
- Proteases
-
Enzymes that break down the peptide bonds that link amino acids together in proteins and polypeptides in a process known as proteolysis; they are also known as peptidases, proteolytic enzymes or proteinases.
- Skin desquamation
-
The physiological peeling or shedding of the outermost corneocytes of the skin. A typical cycle of skin desquamation (which takes ~14 days) involves the apical movement and terminal differentiation of skin keratinocytes into corneocytes and their eventual shedding after cleavage by skin-associated proteases.
- Seminal liquefaction
-
The enzymatic breakdown of the seminal gel — formed by proteins from the seminal vesicles — to become more liquefied. A typical seminal liquefaction cycle is completed within 20 minutes following ejaculation.
- Catalytic triad
-
The three conserved amino-acid residues that are at the centre of the active sites of many enzymes (for example, proteases, amidases, esterases and lipases) and synergistically account for their activity.
- Prime side
-
According to Schechter and Berger's nomenclature of interactions between protease active sites and peptidic or protein substrates, enzyme binding subsites (S′) and their corresponding substrate peptide (P′) residues that are C-terminal to the scissile peptide bond are termed 'prime side' and designated as S1′, S2′ and P1′, P2′, respectively.
- Phage display
-
A molecular laboratory technique for the production and screening of novel proteins or polypeptides. The desired gene fragment is expressed by bacteriophages, which display the resulting protein on their surface where it can be tested for biological activity or interactions with other proteins, peptides or DNA.
- Lamellar granules
-
Specialized secretory organelles typically found in keratinocytes and type 2 pneumocytes that are also known as membrane-coating granules, lamellar bodies, keratinosomes or Odland bodies. The keratinocyte trans-Golgi network and lamellar granules are parts of the same continuous membrane structure, in which lamellar granules transport their cargo and fuse with the cell membrane at the border of the stratum granulosum and stratum corneum to release their contents into the extracellular milieu.
- Acanthosis
-
An abnormal, diffuse hypertrophy of the stratum spinosum, as in eczema and psoriasis.
- Long-term potentiation
-
(LTP) The biological process behind the persistent increase and long-lasting enhancement in synaptic strength following electrical or chemical stimulation of neurons. It is a crucial cellular mechanism for both learning memory (also known as long-term memory) and working memory (also known as short-term memory).
- Chondromyces crocatus
-
A Gram-negative bacteria strain that belongs to the myxobacteria family. They live predominantly in the soil and feed on insoluble organic substances. Although they are poorly distributed ecologically, numerous secondary metabolite products of Chondromyces crocatus have been recently found.
Rights and permissions
About this article
Cite this article
Prassas, I., Eissa, A., Poda, G. et al. Unleashing the therapeutic potential of human kallikrein-related serine proteases. Nat Rev Drug Discov 14, 183–202 (2015). https://doi.org/10.1038/nrd4534
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrd4534
This article is cited by
-
Remodelling of the tumour microenvironment by the kallikrein-related peptidases
Nature Reviews Cancer (2022)
-
Intracellular MUC20 variant 2 maintains mitochondrial calcium homeostasis and enhances drug resistance in gastric cancer
Gastric Cancer (2022)
-
A KLK4 proteinase substrate capture approach to antagonize PAR1
Scientific Reports (2021)
-
Substrate-biased activity-based probes identify proteases that cleave receptor CDCP1
Nature Chemical Biology (2021)
-
Prognostic value of kallikrein-related peptidase 7 (KLK7) mRNA expression in advanced high-grade serous ovarian cancer
Journal of Ovarian Research (2020)