Key Points
-
Notch signalling has key roles in the development and homeostasis of most organs, including — notably — the haematopoietic system, skin, vascular system and liver.
-
An increasing number of known human diseases relate to dysregulated Notch signalling, including developmental congenital disorders and cancers with de novo NOTCH mutations.
-
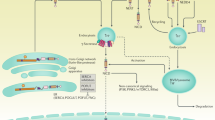
The core pathway is simple but provides ample potential therapeutic possibilities. Viable targets include inhibition of receptor cleavage by γ-secretase inhibitors and blockade of specific receptors or ligands with antibodies.
-
Crosstalk between Notch and other signalling mechanisms may provide possibilities for combinatorial treatments, especially in cancer where targeting several pathways simultaneously may offer considerable benefits. Numerous clinical trials are underway with the aim of modulating Notch signalling.
Abstract
The Notch signalling pathway is evolutionarily conserved and is crucial for the development and homeostasis of most tissues. Deregulated Notch signalling leads to various diseases, such as T cell leukaemia, Alagille syndrome and a stroke and dementia syndrome known as CADASIL, and so strategies to therapeutically modulate Notch signalling are of interest. Clinical trials of Notch pathway inhibitors in patients with solid tumours have been reported, and several approaches are under preclinical evaluation. In this Review, we focus on aspects of the pathway that are amenable to therapeutic intervention, diseases that could be targeted and the various Notch pathway modulation strategies that are currently being explored.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Ellisen, L. W. et al. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell 66, 649–661 (1991). This report describes the first characterization of a mammalian Notch gene and the first NOTCH1 translocations in T-ALL.
Joutel, A. et al. Notch3 mutations in CADASIL, a hereditary adult-onset condition causing stroke and dementia. Nature 383, 707–710 (1996). This study identifies NOTCH3 as the gene that is mutated in the vascular cerebral stroke disorder CADASIL, which was the first finding of NOTCH mutations in a hereditary disorder.
Oda, T. et al. Mutations in the human Jagged1 gene are responsible for Alagille syndrome. Nature Genet. 16, 235–242 (1997).
McDaniell, R. et al. NOTCH2 mutations cause Alagille syndrome, a heterogeneous disorder of the notch signaling pathway. Am. J. Hum. Genet. 79, 169–173 (2006).
Puente, X. S. et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature 475, 101–105 (2011).
Fabbri, G. et al. Analysis of the chronic lymphocytic leukemia coding genome: role of NOTCH1 mutational activation. J. Exp. Med. 208, 1389–1401 (2011).
Sparrow, D. B., Guillén-Navarro, E., Fatkin, D. & Dunwoodie, S. L. Mutation of Hairy-and-Enhancer-of-Split-7 in humans causes spondylocostal dysostosis. Hum. Mol. Genet. 17, 3761–3766 (2008).
Sparrow, D. B. et al. Mutation of the LUNATIC FRINGE gene in humans causes spondylocostal dysostosis with a severe vertebral phenotype. Am. J. Hum. Genet. 78, 28–37 (2006).
Whittock, N. V. et al. Mutated MESP2 causes spondylocostal dysostosis in humans. Am. J. Hum. Genet. 74, 1249–1254 (2004).
Andersson, E. R., Sandberg, R. & Lendahl, U. Notch signaling: simplicity in design, versatility in function. Development 138, 3593–3612 (2011).
Guruharsha, K. G., Kankel, M. W. & Artavanis-Tsakonas, S. The Notch signalling system: recent insights into the complexity of a conserved pathway. Nature Rev. Genet. 13, 654–666 (2012).
Shao, H., Huang, Q. & Liu, Z.-J. Targeting Notch signaling for cancer therapeutic intervention. Adv. Pharmacol. 65, 191–234 (2012).
Groth, C. & Fortini, M. E. Therapeutic approaches to modulating Notch signaling: current challenges and future prospects. Semin. Cell Dev. Biol. 23, 465–472 (2012).
Bray, S. J. Notch signalling: a simple pathway becomes complex. Nature Rev. Mol. Cell. Biol. 7, 678–689 (2006).
Coleman, M. L. et al. Asparaginyl hydroxylation of the Notch ankyrin repeat domain by factor inhibiting hypoxia-inducible factor. J. Biol. Chem. 282, 24027–24038 (2007).
Zheng, X. et al. Interaction with factor inhibiting HIF-1 defines an additional mode of cross-coupling between the Notch and hypoxia signaling pathways. Proc. Natl Acad. Sci. USA 105, 3368–3373 (2008).
Espinosa, L., Ingles-Esteve, J., Aguilera, C. & Bigas, A. Phosphorylation by glycogen synthase kinase-3β down-regulates Notch activity, a link for Notch and Wnt pathways. J. Biol. Chem. 278, 32227–32235 (2003).
King, B. et al. The ubiquitin ligase FBXW7 modulates leukemia-initiating cell activity by regulating MYC stability. Cell 153, 1552–1566 (2013).
Richards, G. S. & Degnan, B. M. The dawn of developmental signaling in the metazoa. Cold Spring Harb. Symp. Quant. Biol. 74, 81–90 (2009).
Ladi, E. et al. The divergent DSL ligand Dll3 does not activate Notch signaling but cell autonomously attenuates signaling induced by other DSL ligands. J. Cell Biol. 170, 983–992 (2005).
Geffers, I. et al. Divergent functions and distinct localization of the Notch ligands DLL1 and DLL3 in vivo. J. Cell Biol. 178, 465–476 (2007).
Yamamoto, S. et al. A mutation in EGF repeat-8 of Notch discriminates between Serrate/Jagged and Delta family ligands. Science 338, 1229–1232 (2012).
Andrawes, M. B. et al. Intrinsic selectivity of Notch 1 for Delta-like 4 over Delta-like 1. J. Biol. Chem. 288, 25477–25489 (2013).
Van de Walle, I. et al. Specific Notch receptor-ligand interactions control human TCR-αβ/γδ development by inducing differential Notch signal strength. J. Exp. Med. 210, 683–697 (2013).
Panin, V. M., Papayannopoulos, V., Wilson, R. & Irvine, K. D. Fringe modulates Notch-ligand interactions. Nature 387, 908–912 (1997).
Hicks, C. et al. Fringe differentially modulates Jagged1 and Delta1 signalling through Notch1 and Notch2. Nature Cell Biol. 2, 515–520 (2000).
Rana, N. A. & Haltiwanger, R. S. Fringe benefits: functional and structural impacts of O-glycosylation on the extracellular domain of Notch receptors. Curr. Opin. Struct. Biol. 21, 583–589 (2011).
Jin, S. et al. Non-canonical Notch signaling activates IL-6/JAK/STAT signaling in breast tumor cells and is controlled by p53 and IKKα/IKKβ. Oncogene 32, 4892–4902 (2012).
Shimizu, K. et al. Mouse Jagged1 physically interacts with Notch2 and other Notch receptors. Assessment by quantitative methods. J. Biol. Chem. 274, 32961–32969 (1999).
Rebay, I. et al. Specific EGF repeats of Notch mediate interactions with Delta and Serrate: implications for Notch as a multifunctional receptor. Cell 67, 687–699 (1991).
Falk, R. et al. Generation of anti-Notch antibodies and their application in blocking Notch signalling in neural stem cells. Methods 58, 69–78 (2012).
Liu, S. K. et al. Delta-like ligand 4-Notch blockade and tumor radiation response. J. Natl Cancer Inst. 103, 1–21 (2011).
Ohishi, K., Varnum-Finney, B., Serda, R. E., Anasetti, C. & Bernstein, I. D. The Notch ligand, Delta-1, inhibits the differentiation of monocytes into macrophages but permits their differentiation into dendritic cells. Blood 98, 1402–1407 (2001).
Ohishi, K., Varnum-Finney, B. & Bernstein, I. D. Delta-1 enhances marrow and thymus repopulating ability of human CD34+CD38− cord blood cells. J. Clin. Invest. 110, 1165–1174 (2002).
Ohishi, K. et al. Monocytes express high amounts of Notch and undergo cytokine specific apoptosis following interaction with the Notch ligand, Delta-1. Blood 95, 2847–2854 (2000).
Hicks, C. et al. A secreted Delta1-Fc fusion protein functions both as an activator and inhibitor of Notch1 signaling. J. Neurosci. Res. 68, 655–667 (2002).
Czajkowsky, D. M., Hu, J., Shao, Z. & Pleass, R. J. Fc-fusion proteins: new developments and future perspectives. EMBO Mol. Med. 4, 1015–1028 (2012).
Gonçalves, R. M., Martins, M. C. L., Almeida-Porada, G. & Barbosa, M. A. Induction of notch signaling by immobilization of jagged-1 on self-assembled monolayers. Biomaterials 30, 6879–6887 (2009).
Toda, H., Yamamoto, M., Kohara, H. & Tabata, Y. Orientation-regulated immobilization of Jagged1 on glass substrates for ex vivo proliferation of a bone marrow cell population containing hematopoietic stem cells. Biomaterials 32, 6920–6928 (2011).
Varnum-Finney, B., Brashem-Stein, C. & Bernstein, I. D. Combined effects of Notch signaling and cytokines induce a multiple log increase in precursors with lymphoid and myeloid reconstituting ability. Blood 101, 1784–1789 (2003).
Conboy, I. M., Conboy, M. J., Smythe, G. M. & Rando, T. A. Notch-mediated restoration of regenerative potential to aged muscle. Science 302, 1575–1577 (2003).
Small, D. et al. Soluble Jagged 1 represses the function of its transmembrane form to induce the formation of the Src-dependent chord-like phenotype. J. Biol. Chem. 276, 32022–32030 (2001).
Trifonova, R. et al. The non-transmembrane form of Delta1, but not of Jagged1, induces normal migratory behavior accompanied by fibroblast growth factor receptor 1-dependent transformation. J. Biol. Chem. 279, 13285–13288 (2004).
Li, L. et al. The human homolog of rat Jagged1 expressed by marrow stroma inhibits differentiation of 32D cells through interaction with Notch1. Immunity 8, 43–55 (1998).
Kannan, S. et al. Notch activation inhibits AML growth and survival: a potential therapeutic approach. J. Exp. Med. 210, 321–337 (2013).
Androutsellis-Theotokis, A. et al. Notch signalling regulates stem cell numbers in vitro and in vivo. Nature 442, 823–826 (2006).
Lu, J. et al. Endothelial cells promote the colorectal cancer stem cell phenotype through a soluble form of Jagged-1. Cancer Cell 23, 171–185 (2013).
Nehring, L. C., Miyamoto, A., Hein, P. W., Weinmaster, G. & Shipley, J. M. The extracellular matrix protein MAGP-2 interacts with Jagged1 and induces its shedding from the cell surface. J. Biol. Chem. 280, 20349–20355 (2005).
Chillakuri, C. R. et al. Structural analysis uncovers lipid-binding properties of Notch ligands. Cell Rep. 5, 861–867 (2013).
Cordle, J. et al. A conserved face of the Jagged/Serrate DSL domain is involved in Notch trans-activation and cis-inhibition. Nature Struct. Mol. Biol. 15, 849–857 (2008).
Logeat, F. et al. The Notch1 receptor is cleaved constitutively by a furin-like convertase. Proc. Natl Acad. Sci. USA 95, 8108–8112 (1998).
Gordon, W. R. et al. Effects of S1 cleavage on the structure, surface export, and signaling activity of human Notch1 and Notch2. PLoS ONE 4, e6613 (2009).
Bush, G. et al. Ligand-induced signaling in the absence of furin processing of Notch1. Dev. Biol. 229, 494–502 (2001).
Kidd, S. & Lieber, T. Furin cleavage is not a requirement for Drosophila Notch function. Mech. Dev. 115, 41–51 (2002).
Seidah, N. G. & Prat, A. The biology and therapeutic targeting of the proprotein convertases. Nature Rev. Drug Discov. 11, 367–383 (2012).
Fernandez-Valdivia, R. et al. Regulation of mammalian Notch signaling and embryonic development by the protein O-glucosyltransferase Rumi. Development 138, 1925–1934 (2011).
Acar, M. et al. Rumi is a CAP10 domain glycosyltransferase that modifies Notch and is required for Notch signaling. Cell 132, 247–258 (2008).
Shi, S. & Stanley, P. Protein O-fucosyltransferase 1 is an essential component of Notch signaling pathways. Proc. Natl Acad. Sci. USA 100, 5234–5239 (2003).
Sethi, M. K. et al. Identification of glycosyltransferase 8 family members as xylosyltransferases acting on O-glucosylated Notch epidermal growth factor repeats. J. Biol. Chem. 285, 1582–1586 (2010).
Goode, S. & Perrimon, N. Brainiac and Fringe are similar pioneer proteins that impart specificity to notch signaling during Drosophila development. Cold Spring Harb. Symp. Quant. Biol. 62, 177–184 (1997).
Moloney, D. J. et al. Fringe is a glycosyltransferase that modifies Notch. Nature 406, 369–375 (2000).
Brückner, K., Perez, L., Clausen, H. & Cohen, S. Glycosyltransferase activity of Fringe modulates Notch–Delta interactions. Nature 406, 411–415 (2000).
Wang, X. & Ha, T. Defining single molecular forces required to activate integrin and notch signaling. Science 340, 991–994 (2013).
Hansson, E. M. et al. Control of Notch-ligand endocytosis by ligand–receptor interaction. J. Cell Sci. 123, 2931–2942 (2010).
Nichols, J. T. et al. DSL ligand endocytosis physically dissociates Notch1 heterodimers before activating proteolysis can occur. J. Cell Biol. 176, 445–458 (2007).
Meloty-Kapella, L. et al. Optical tweezers studies on Notch: single-molecule interaction strength is independent of ligand endocytosis. Dev. Cell 22, 1313–1320 (2012).
Brou, C. et al. A novel proteolytic cleavage involved in Notch signaling: the role of the disintegrin-metalloprotease TACE. Mol. Cell 5, 207–216 (2000).
Stephenson, N. L. & Avis, J. M. Direct observation of proteolytic cleavage at the S2 site upon forced unfolding of the Notch negative regulatory region. Proc. Natl Acad. Sci. USA 109, E2757–E2765 (2012).
Parks, A. L., Klueg, K. M., Stout, J. R. & Muskavitch, M. A. Ligand endocytosis drives receptor dissociation and activation in the Notch pathway. Development 127, 1373–1385 (2000).
Mumm, J. S. et al. A ligand-induced extracellular cleavage regulates γ-secretase-like proteolytic activation of Notch1. Mol. Cell 5, 197–206 (2000).
Van Tetering, G. & Vooijs, M. Proteolytic cleavage of Notch: “HIT and RUN''. Curr. Mol. Med. 11, 255–269 (2011).
Cousin, H., Abbruzzese, G., Kerdavid, E., Gaultier, A. & Alfandari, D. Translocation of the cytoplasmic domain of ADAM13 to the nucleus is essential for calpain8-a expression and cranial neural crest cell migration. Dev. Cell 20, 256–263 (2011).
Tousseyn, T. et al. ADAM10, the rate-limiting protease of regulated intramembrane proteolysis of Notch and other proteins, is processed by ADAMS-9, ADAMS-15, and the γ-secretase. J. Biol. Chem. 284, 11738–11747 (2009).
Wu, Y. et al. Therapeutic antibody targeting of individual Notch receptors. Nature 464, 1052–1057 (2010). This paper reports on the generation and characterization of antibodies that are specific for the NRRs of NOTCH1 and NOTCH2 , thus inhibiting the activation of the receptor and blocking Notch-driven tumour growth.
De Strooper, B. et al. A presenilin-1-dependent γ-secretase-like protease mediates release of Notch intracellular domain. Nature 398, 518–522 (1999).
Struhl, G. & Greenwald, I. Presenilin is required for activity and nuclear access of Notch in Drosophila. Nature 398, 522–525 (1999). References 75 and 76 provide the first link between Notch receptor processing and the γ-secretase complex, a finding that is of considerable importance for research into GSIs and Alzheimer's disease.
Schroeter, E. H., Kisslinger, J. A. & Kopan, R. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature 393, 382–386 (1998).
Gupta-Rossi, N. et al. Monoubiquitination and endocytosis direct γ-secretase cleavage of activated Notch receptor. J. Cell Biol. 166, 73–83 (2004).
Sorensen, E. B. & Conner, S. γ-secretase-dependent cleavage initiates Notch signaling from the plasma membrane. Traffic 11, 1234–1245 (2010).
Tagami, S. et al. Regulation of Notch signaling by dynamic changes in the precision of S3 cleavage of Notch-1. Mol. Cell. Biol. 28, 165–176 (2008).
Vaccari, T., Lu, H., Kanwar, R., Fortini, M. E. & Bilder, D. Endosomal entry regulates Notch receptor activation in Drosophila melanogaster. J. Cell Biol. 180, 755–762 (2008).
Okochi, M. et al. Presenilins mediate a dual intramembranous γ-secretase cleavage of Notch-1. EMBO J. 21, 5408–5416 (2002).
Okochi, M. et al. Secretion of the Notch-1 Aβ-like peptide during Notch signaling. J. Biol. Chem. 281, 7890–7898 (2006).
Krejcí, A. & Bray, S. Notch activation stimulates transient and selective binding of Su(H)/CSL to target enhancers. Genes Dev. 21, 1322–1327 (2007).
Castel, D. et al. Dynamic binding of RBPJ is determined by Notch signaling status. Genes Dev. 27, 1059–1071 (2013).
Wilson, J. J. & Kovall, R. A. Crystal structure of the CSL-Notch-Mastermind ternary complex bound to DNA. Cell 124, 985–996 (2006).
Nam, Y., Sliz, P., Song, L., Aster, J. C. & Blacklow, S. C. Structural basis for cooperativity in recruitment of MAML coactivators to Notch transcription complexes. Cell 124, 973–983 (2006). References 86 and 87 report on the crystal structure of the Notch transcriptional activation complex, including the ankyrin domain of Notch 1 and CSL on DNA, together with a MAML1 polypeptide. This shows that CSL and ankyrin form a groove for MAML binding, and this information is essential for constructing NICD–CSL-inhibiting peptides.
Weng, A. P. et al. Growth suppression of pre-T acute lymphoblastic leukemia cells by inhibition of Notch signaling. Mol. Cell. Biol. 23, 655–664 (2003).
Maillard, I. et al. Mastermind critically regulates Notch-mediated lymphoid cell fate decisions. Blood 104, 1696–1702 (2004).
Maillard, I. et al. The requirement for Notch signaling at the β-selection checkpoint in vivo is absolute and independent of the pre–T cell receptor. J. Exp. Med. 203, 2239–2245 (2006).
Maillard, I. et al. Canonical Notch signaling is dispensable for the maintenance of adult hematopoietic stem cells. Cell Stem Cell 2, 356–366 (2008).
Osanyingbemi-Obidi, J., Dobromilskaya, I., Illei, P. B., Hann, C. L. & Rudin, C. M. Notch signaling contributes to lung cancer clonogenic capacity in vitro but may be circumvented in tumorigenesis in vivo. Mol. Cancer Res. 9, 1746–1754 (2011).
Shen, H. et al. The Notch coactivator, MAML1, functions as a novel coactivator for MEF2C-mediated transcription and is required for normal myogenesis. Genes Dev. 20, 675–688 (2006).
Saint Just Ribeiro, M. & Wallberg, A. E. Transcriptional mechanisms by the coregulator MAML1. Curr. Protein Pept. Sci. 10, 570–576 (2009).
McElhinny, A. S., Li, J.-L. & Wu, L. Mastermind-like transcriptional co-activators: emerging roles in regulating cross talk among multiple signaling pathways. Oncogene 27, 5138–5147 (2008).
Moellering, R. E. et al. Direct inhibition of the NOTCH transcription factor complex. Nature 462, 182–188 (2009).
Yuan, Z., Friedmann, D. R., VanderWielen, B. D., Collins, K. J. & Kovall, R. A. Characterization of CSL (CBF-1, Su(H), Lag-1) mutants reveals differences in signaling mediated by Notch1 and Notch2. J. Biol. Chem. 287, 34904–34916 (2012).
Lubman, O. Y., Ilagan, M. X. G., Kopan, R. & Barrick, D. Quantitative dissection of the Notch:CSL interaction: insights into the Notch-mediated transcriptional switch. J. Mol. Biol. 365, 577–589 (2007).
Johnson, J. E. & Macdonald, R. J. Notch-independent functions of CSL. Curr. Top. Dev. Biol. 97, 55–74 (2011).
Foltz, D. R., Santiago, M. C., Berechid, B. E. & Nye, J. S. Glycogen synthase kinase-3β modulates Notch signaling and stability. Curr. Biol. 12, 1006–1011 (2002).
Fryer, C. J., White, J. B. & Jones, K. A. Mastermind recruits CycC:CDK8 to phosphorylate the Notch ICD and coordinate activation with turnover. Mol. Cell 16, 509–520 (2004).
Sjöqvist, M. et al. PKCζ regulates Notch receptor routing and activity in a Notch signaling-dependent manner. Cell Res. http://dx.doi.org/10.1038/cr.2014.34 (2014).
Guarani, V. et al. Acetylation-dependent regulation of endothelial Notch signalling by the SIRT1 deacetylase. Nature 473, 234–238 (2011).
Le Bras, S., Loyer, N. & Le Borgne, R. The multiple facets of ubiquitination in the regulation of Notch signaling pathway. Traffic 12, 149–161 (2011).
Hubbard, E. J. A., Wu, G., Kitajewski, J. & Greenwald, I. sel-10, a negative regulator of lin-12 activity in Caenorhabditis elegans, encodes a member of the CDC4 family of proteins. Genes Dev. 11, 3182–3193 (1997).
Tsunematsu, R. et al. Mouse Fbw7/Sel-10/Cdc4 is required for Notch degradation during vascular development. J. Biol. Chem. 279, 9417–9423 (2004).
Malyukova, A. et al. The tumor suppressor gene hCDC4 is frequently mutated in human T-cell acute lymphoblastic leukemia with functional consequences for Notch signaling. Cancer Res. 67, 5611–5616 (2007).
O'Neil, J. et al. FBW7 mutations in leukemic cells mediate NOTCH pathway activation and resistance to γ-secretase inhibitors. J. Exp. Med. 204, 1813–1824 (2007).
McGill, M. A. & McGlade, C. J. Mammalian Numb proteins promote Notch1 receptor ubiquitination and degradation of the Notch1 intracellular domain. J. Biol. Chem. 278, 23196–23203 (2003).
Qiu, L. et al. Recognition and ubiquitination of Notch by Itch, a hect-type E3 ubiquitin ligase. J. Biol. Chem. 275, 35734–35737 (2000).
Pettersson, S. et al. Non-degradative ubiquitination of the Notch1 receptor by the E3 ligase MDM2 activates the Notch signalling pathway. Biochem. J. 450, 523–536 (2013).
Zhang, J., Liu, M., Su, Y., Du, J. & Zhu, A. J. A targeted in vivo RNAi screen reveals deubiquitinases as new regulators of Notch signaling. G3 2, 1563–1575 (2012).
Muñoz-Descalzo, S., Tkocz, K., Balayo, T. & Arias, A. M. Modulation of the ligand-independent traffic of Notch by Axin and Apc contributes to the activation of Armadillo in Drosophila. Development 138, 1501–1506 (2011).
Hayward, P., Balayo, T. & Martinez Arias, A. Notch synergizes with axin to regulate the activity of armadillo in Drosophila. Dev. Dyn. 235, 2656–2666 (2006).
Hayward, P. et al. Notch modulates Wnt signalling by associating with Armadillo/β-catenin and regulating its transcriptional activity. Development 132, 1819–1830 (2005).
Shimizu, T. et al. Stabilized β-catenin functions through TCF/LEF proteins and the Notch/RBP-Jκ complex to promote proliferation and suppress differentiation of neural precursor cells. Mol. Cell. Biol. 28, 7427–7441 (2008).
Yamamizu, K. et al. Convergence of Notch and β-catenin signaling induces arterial fate in vascular progenitors. J. Cell Biol. 189, 325–338 (2010).
Axelrod, J. D., Matsuno, K., Artavanis-Tsakonas, S. & Perrimon, N. Interaction between Wingless and Notch signaling pathways mediated by dishevelled. Science 271, 1826–1832 (1996).
Muñoz-Descalzo, S. et al. Wingless modulates the ligand independent traffic of Notch through Dishevelled. Fly 4, 182–193 (2010).
Blokzijl, A. et al. Cross-talk between the Notch and TGF-β signaling pathways mediated by interaction of the Notch intracellular domain with Smad3. J. Cell Biol. 163, 723–728 (2003).
Dahlqvist, C. et al. Functional Notch signaling is required for BMP4-induced inhibition of myogenic differentiation. Development 130, 6089–6099 (2003).
Itoh, F. Synergy and antagonism between Notch and BMP receptor signaling pathways in endothelial cells. EMBO J. 23, 541–551 (2004).
Gustafsson, M. V. et al. Hypoxia requires Notch signaling to maintain the undifferentiated cell state. Dev. Cell 9, 617–628 (2005). This is the first report demonstrating crosstalk between Notch signalling and the cellular hypoxic response, which is important when considering therapeutic modulation of the cellular hypoxic response.
Roti, G. et al. Complementary genomic screens identify SERCA as a therapeutic target in NOTCH1 mutated cancer. Cancer Cell 23, 390–405 (2013).
Ilagan, M. X. G. & Kopan, R. Selective blockade of transport via SERCA inhibition: the answer for oncogenic forms of Notch? Cancer Cell 23, 267–269 (2013).
Andersen, P., Uosaki, H., Shenje, L. T. & Kwon, C. Non-canonical Notch signaling: emerging role and mechanism. Trends Cell Biol. 22, 257–265 (2012).
D'Souza, B., Meloty-Kapella, L. & Weinmaster, G. Canonical and non-canonical Notch ligands. Curr. Top. Dev. Biol. 92, 73–129 (2010).
Sandy, A. R., Jones, M. & Maillard, I. Notch signaling and development of the hematopoietic system. Adv. Exp. Med. Biol. 727, 71–88 (2012).
Xu, K., Moghal, N. & Egan, S. E. Notch signaling in lung development and disease. Adv. Exp. Med. Biol. 727, 89–98 (2012).
Barak, H., Surendran, K. & Boyle, S. C. The role of Notch signaling in kidney development and disease. Adv. Exp. Med. Biol. 727, 99–113 (2012).
Kume, T. Ligand-dependent Notch signaling in vascular formation. Adv. Exp. Med. Biol. 727, 210–222 (2012).
Murata, J., Ikeda, K. & Okano, H. Notch signaling and the developing inner ear. Adv. Exp. Med. Biol. 727, 161–173 (2012).
Aubin-Houzelstein, G. Notch signaling and the developing hair follicle. Adv. Exp. Med. Biol. 727, 142–160 (2012).
Mead, T. J. & Yutzey, K. E. Notch signaling and the developing skeleton. Adv. Exp. Med. Biol. 727, 114–130 (2012).
Clements, W. K. et al. A somitic Wnt16/Notch pathway specifies haematopoietic stem cells. Nature 474, 220–224 (2011).
Han, H. et al. Inducible gene knockout of transcription factor recombination signal binding protein-J reveals its essential role in T versus B lineage decision. Int. Immunol. 14, 637–645 (2002).
Tanigaki, K. et al. Notch-RBP-J signaling is involved in cell fate determination of marginal zone B cells. Nature Immunol. 3, 443–450 (2002).
Ciofani, M. & Zúñiga-Pflücker, J. C. Notch promotes survival of pre-T cells at the β-selection checkpoint by regulating cellular metabolism. Nature Immunol. 6, 881–888 (2005).
Lobry, C. et al. Notch pathway activation targets AML-initiating cell homeostasis and differentiation. J. Exp. Med. 210, 301–319 (2013).
Schroeder, T. & Just, U. Notch signalling via RBP-J promotes myeloid differentiation. EMBO J. 19, 2558–2568 (2000).
Bigas, A., Martin, D. I. & Milner, L. A. Notch1 and Notch2 inhibit myeloid differentiation in response to different cytokines. Mol. Cell. Biol. 18, 2324–2333 (1998).
Oh, P. et al. In vivo mapping of notch pathway activity in normal and stress hematopoiesis. Cell Stem Cell 13, 190–204 (2013).
Klinakis, A. et al. A novel tumour-suppressor function for the Notch pathway in myeloid leukaemia. Nature 473, 230–233 (2011).
Li, Y.-P. et al. Human mesenchymal stem cells license adult CD34+ hemopoietic progenitor cells to differentiate into regulatory dendritic cells through activation of the Notch pathway. J. Immunol. 180, 1598–1608 (2008).
Bigas, A., D'Altri, T. & Espinosa, L. The Notch pathway in hematopoietic stem cells. Curr. Top. Microbiol. Immunol. 360, 1–18 (2012).
Weng, A. P. et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science 306, 269–271 (2004). This study reports that over 50% of T-ALL cases contain NOTCH -activating mutations, as a result of a mutation in the PEST domain or in the heterodimerization domain, which lead to hyperactivation.
Thompson, B. J. et al. The SCFFBW7 ubiquitin ligase complex as a tumor suppressor in T cell leukemia. J. Exp. Med. 204, 1825–1835 (2007).
Henkel, T., Ling, P. D., Hayward, S. D. & Peterson, M. G. Mediation of Epstein–Barr virus EBNA2 transactivation by recombination signal-binding protein Jκ. Science 265, 92–95 (1994).
Cotter, M., Callahan, J., Aster, J. & Robertson, E. Intracellular forms of human NOTCH1 functionally activate essential Epstein–Barr virus major latent promoters in the Burkitt's lymphoma BJAB cell line but repress these promoters in Jurkat cells. J. Virol. 74, 1486–1494 (2000).
Strobl, L. J. et al. Activated Notch1 modulates gene expression in B cells similarly to Epstein-Barr viral nuclear antigen 2. J. Virol. 74, 1727–1735 (2000).
Callahan, J., Aster, J., Sklar, J., Kieff, E. & Robertson, E. S. Intracellular forms of human NOTCH1 interact at distinctly different levels with RBP-jkappa in human B and T cells. Leukemia 14, 84–92 (2000).
Kridel, R. et al. Whole transcriptome sequencing reveals recurrent NOTCH1 mutations in mantle cell lymphoma. Blood 119, 1963–1971 (2012).
Dumortier, A. et al. Atopic dermatitis-like disease and associated lethal myeloproliferative disorder arise from loss of Notch signaling in the murine skin. PLoS ONE 5, e9258 (2010).
The Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 368, 2059–2074 (2013).
Grieselhuber, N. R. et al. Notch signaling in acute promyelocytic leukemia. Leukemia 27, 1548–1557 (2013).
Wang, Q., Zhao, N., Kennard, S. & Lilly, B. Notch2 and Notch3 function together to regulate vascular smooth muscle development. PLoS ONE 7, e37365 (2012).
Domenga, V. et al. Notch3 is required for arterial identity and maturation of vascular smooth muscle cells. Genes Dev. 18, 2730–2735 (2004).
Liu, H., Zhang, W., Kennard, S., Caldwell, R. B. & Lilly, B. Notch3 is critical for proper angiogenesis and mural cell investment. Circ. Res. 107, 860–870 (2010).
Oka, C. et al. Disruption of the mouse RBP-J κ gene results in early embryonic death. Development 121, 3291–3301 (1995).
Gale, N. W. et al. Haploinsufficiency of delta-like 4 ligand results in embryonic lethality due to major defects in arterial and vascular development. Proc. Natl Acad. Sci. USA 101, 15949–15954 (2004).
Xue, Y. et al. Embryonic lethality and vascular defects in mice lacking the Notch ligand Jagged1. Hum. Mol. Genet. 8, 723–730 (1999).
McCright, B. et al. Defects in development of the kidney, heart and eye vasculature in mice homozygous for a hypomorphic Notch2 mutation. Development 128, 491–502 (2001).
Krebs, L. T. et al. Notch signaling is essential for vascular morphogenesis in mice. Genes Dev. 14, 1343–1352 (2000).
Lawson, N. D., Vogel, A. M. & Weinstein, B. M. Sonic hedgehog and vascular endothelial growth factor act upstream of the Notch pathway during arterial endothelial differentiation. Dev. Cell 3, 127–136 (2002).
Zhong, T. P., Childs, S., Leu, J. P. & Fishman, M. C. Gridlock signalling pathway fashions the first embryonic artery. Nature 414, 216–220 (2001).
Jakobsson, L. et al. Endothelial cells dynamically compete for the tip cell position during angiogenic sprouting. Nature Cell Biol. 12, 943–953 (2010).
Liu, Z. et al. Notch1 loss of heterozygosity causes vascular tumors and lethal hemorrhage in mice. J. Clin. Invest. 121, 800–808 (2011).
Yan, M. et al. Chronic DLL4 blockade induces vascular neoplasms. Nature 463, E6–E7 (2010). This paper demonstrates that long-term use of DLL4 antibodies and DLL4-antagonistic peptides induces endothelial cell activation and bile duct proliferation, as well as vascular tumours, in rats and monkeys. This suggests that although this therapy is more directly targeted than broad-spectrum Notch inhibition, it poses risks.
Mosquera, J.-M. et al. Novel MIR143-NOTCH fusions in benign and malignant glomus tumors. Genes. Chromosomes Cancer 52, 1075–1087 (2013).
Joutel, A. et al. Strong clustering and stereotyped nature of Notch3 mutations in CADASIL patients. Lancet 350, 1511–1515 (1997).
Monet-Leprêtre, M. et al. Abnormal recruitment of extracellular matrix proteins by excess Notch3 ECD: a new pathomechanism in CADASIL. Brain 136, 1830–1845 (2013).
Joutel, A. et al. Cerebrovascular dysfunction and microcirculation rarefaction precede white matter lesions in a mouse genetic model of cerebral ischemic small vessel disease. J. Clin. Invest. 120, 433–445 (2010).
Li, X. et al. Notch3 signaling promotes the development of pulmonary arterial hypertension. Nature Med. 15, 1289–1297 (2009). This study shows that Notch 3 is upregulated in human PAH in small pulmonary artery smooth muscle cells and that disease severity correlates with Notch 3 protein levels in the lung. In addition, Notch 3 activity is required for the development of PAH in a mouse model of this disease.
Yu, Y.-R. A., Mao, L., Piantadosi, C. A. & Gunn, M. D. CCR2 deficiency, dysregulation of Notch signaling, and spontaneous pulmonary arterial hypertension. Am. J. Respir. Cell. Mol. Biol. 48, 647–654 (2013).
De la Pompa, J. L. & Epstein, J. A. Coordinating tissue interactions: Notch signaling in cardiac development and disease. Dev. Cell 22, 244–254 (2012).
Timmerman, L. A. et al. Notch promotes epithelial-mesenchymal transition during cardiac development and oncogenic transformation. Genes Dev. 18, 99–115 (2004).
Grego-Bessa, J. et al. Notch signaling is essential for ventricular chamber development. Dev. Cell 12, 415–429 (2007).
High, F. A. et al. Murine Jagged1/Notch signaling in the second heart field orchestrates Fgf8 expression and tissue-tissue interactions during outflow tract development. J. Clin. Invest. 119, 1986–1996 (2009).
Jain, R. et al. Cardiac neural crest orchestrates remodeling and functional maturation of mouse semilunar valves. J. Clin. Invest. 121, 422–430 (2011).
Rutenberg, J. B. et al. Developmental patterning of the cardiac atrioventricular canal by Notch and Hairy-related transcription factors. Development 133, 4381–4390 (2006).
Watanabe, Y. et al. Activation of Notch1 signaling in cardiogenic mesoderm induces abnormal heart morphogenesis in mouse. Development 133, 1625–1634 (2006).
Luna-Zurita, L. et al. Integration of a Notch-dependent mesenchymal gene program and Bmp2-driven cell invasiveness regulates murine cardiac valve formation. J. Clin. Invest. 120, 3493–3507 (2010).
Rentschler, S. et al. Notch signaling regulates murine atrioventricular conduction and the formation of accessory pathways. J. Clin. Invest. 121, 525–533 (2011).
Garg, V. et al. Mutations in NOTCH1 cause aortic valve disease. Nature 437, 270–274 (2005).
Mohamed, S. A. et al. Novel missense mutations (p. T596M and p. P1797H) in NOTCH1 in patients with bicuspid aortic valve. Biochem. Biophys. Res. Commun. 345, 1460–1465 (2006).
McKellar, S. H. et al. Novel NOTCH1 mutations in patients with bicuspid aortic valve disease and thoracic aortic aneurysms. J. Thorac. Cardiovasc. Surg. 134, 290–296 (2007).
Nigam, V. & Srivastava, D. Notch1 represses osteogenic pathways in aortic valve cells. J. Mol. Cell. Cardiol. 47, 828–834 (2009).
Nus, M. et al. Diet-induced aortic valve disease in mice haploinsufficient for the Notch pathway effector RBPJK/CSL. Arterioscler. Thromb. Vasc. Biol. 31, 1580–1588 (2011).
Isidor, B. et al. Truncating mutations in the last exon of NOTCH2 cause a rare skeletal disorder with osteoporosis. Nature Genet. 43, 306–308 (2011).
Simpson, M. A. et al. Mutations in NOTCH2 cause Hajdu–Cheney syndrome, a disorder of severe and progressive bone loss. Nature Genet. 43, 303–305 (2011). References 189 and 190 identify nonsense and frameshift mutations in NOTCH2 in patients with Hajdu–Cheney syndrome.
Lowell, S., Jones, P., Le Roux, I., Dunne, J. & Watt, F. M. Stimulation of human epidermal differentiation by Delta–Notch signalling at the boundaries of stem-cell clusters. Curr. Biol. 10, 491–500 (2000).
Thelu, J., Rossio, P., Favier, B. & Thélu, J. Notch signalling is linked to epidermal cell differentiation level in basal cell carcinoma, psoriasis and wound healing. BMC Dermatol. 2, 7 (2002).
Rangarajan, A. et al. Notch signaling is a direct determinant of keratinocyte growth arrest and entry into differentiation. EMBO J. 20, 3427–3436 (2001).
Mammucari, C. et al. Integration of Notch 1 and calcineurin/NFAT signaling pathways in keratinocyte growth and differentiation control. Dev. Cell 8, 665–676 (2005).
Blanpain, C., Lowry, W. E., Pasolli, H. A. & Fuchs, E. Canonical notch signaling functions as a commitment switch in the epidermal lineage. Genes Dev. 20, 3022–3035 (2006).
Nicolas, M. et al. Notch1 functions as a tumor suppressor in mouse skin. Nature Genet. 33, 416–421 (2003). This report provides the first genetic evidence of a role for Notch as a tumour suppressor gene.
Demehri, S. et al. Notch-deficient skin induces a lethal systemic B-lymphoproliferative disorder by secreting TSLP, a sentinel for epidermal integrity. PLoS Biol. 6, e123 (2008).
Di Piazza, M., Nowell, C. S., Koch, U., Durham, A.-D. & Radtke, F. Loss of cutaneous TSLP-dependent immune responses skews the balance of inflammation from tumor protective to tumor promoting. Cancer Cell 22, 479–493 (2012).
Demehri, S. et al. Elevated epidermal thymic stromal lymphopoietin levels establish an antitumor environment in the skin. Cancer Cell 22, 494–505 (2012).
Pickering, C. R. et al. Integrative genomic characterization of oral squamous cell carcinoma identifies frequent somatic drivers. Cancer Discov. 3, 770–781 (2013).
Jithesh, P. V. et al. The epigenetic landscape of oral squamous cell carcinoma. Br. J. Cancer 108, 370–379 (2013).
Yoshida, R. et al. The pathological significance of Notch1 in oral squamous cell carcinoma. Lab. Invest. 93, 1068–1081 (2013).
Pourquié, O. Vertebrate segmentation: from cyclic gene networks to scoliosis. Cell 145, 650–663 (2011).
Dequéant, M.-L. et al. A complex oscillating network of signaling genes underlies the mouse segmentation clock. Science 314, 1595–1598 (2006).
Dias, A. S., de Almeida, I., Belmonte, J. M., Glazier, J. A. & Stern, C. D. Somites without a clock. Science 343, 791–795 (2014).
Bulman, M. P. et al. Mutations in the human Delta homologue, DLL3, cause axial skeletal defects in spondylocostal dysostosis. Nature Genet. 24, 438–441 (2000).
Cornier, A. S. et al. Mutations in the MESP2 gene cause spondylothoracic dysostosis/Jarcho-Levin syndrome. Am. J. Hum. Genet. 82, 1334–1341 (2008).
Sparrow, D. B. et al. A mechanism for gene-environment interaction in the etiology of congenital scoliosis. Cell 149, 295–306 (2012). This report describes a link between an environmental cue (hypoxia) and genetic mutations in the Notch signalling system for disease aetiology.
Lewis, J. Rules for the production of sensory cells. Ciba Found. Symp. 160, 25–39; discussion 40–53 (1991).
Kiernan, A. E., Cordes, R., Kopan, R., Gossler, A. & Gridley, T. The Notch ligands DLL1 and JAG2 act synergistically to regulate hair cell development in the mammalian inner ear. Development 132, 4353–4362 (2005).
Haddon, C., Jiang, Y. J., Smithers, L. & Lewis, J. Delta-Notch signalling and the patterning of sensory cell differentiation in the zebrafish ear: evidence from the mind bomb mutant. Development 125, 4637–4644 (1998).
Haddon, C. et al. Hair cells without supporting cells: further studies in the ear of the zebrafish mind bomb mutant. J. Neurocytol. 28, 837–850 (1999).
Tateya, T., Imayoshi, I., Tateya, I., Ito, J. & Kageyama, R. Cooperative functions of Hes/Hey genes in auditory hair cell and supporting cell development. Dev. Biol. 352, 329–340 (2011).
Kiernan, A. E. et al. The Notch ligand Jagged1 is required for inner ear sensory development. Proc. Natl Acad. Sci. USA 98, 3873–3878 (2001).
Tsai, H. et al. The mouse slalom mutant demonstrates a role for Jagged1 in neuroepithelial patterning in the organ of Corti. Hum. Mol. Genet. 10, 507–512 (2001).
Vrijens, K. et al. Ozzy, a Jag1 vestibular mouse mutant, displays characteristics of Alagille syndrome. Neurobiol. Dis. 24, 28–40 (2006).
McCright, B., Lozier, J. & Gridley, T. A mouse model of Alagille syndrome: Notch2 as a genetic modifier of Jag1 haploinsufficiency. Development 129, 1075–1082 (2002).
Yanger, K. et al. Robust cellular reprogramming occurs spontaneously during liver regeneration. Genes Dev. 27, 719–724 (2013).
Pajvani, U. B. et al. Inhibition of Notch uncouples Akt activation from hepatic lipid accumulation by decreasing mTorc1 stability. Nature Med. 19, 1054–1060 (2013).
Zender, S. et al. A critical role for Notch signaling in the formation of cholangiocellular carcinomas. Cancer Cell 23, 784–795 (2013).
Villanueva, A. et al. Notch signaling is activated in human hepatocellular carcinoma and induces tumor formation in mice. Gastroenterology 143, 1660–1669.e7 (2012). This paper shows that NICD produces hepatocellular carcinoma in mice and reveals a Notch expression signature in patients with hepatocellular carcinoma.
Raouf, A. et al. Transcriptome analysis of the normal human mammary cell commitment and differentiation process. Cell Stem Cell 3, 109–118 (2008).
Bouras, T. et al. Notch signaling regulates mammary stem cell function and luminal cell-fate commitment. Cell Stem Cell 3, 429–441 (2008).
Pece, S. et al. Loss of negative regulation by Numb over Notch is relevant to human breast carcinogenesis. J. Cell Biol. 167, 215–221 (2004).
Stylianou, S., Clarke, R. B. & Brennan, K. Aberrant activation of Notch signaling in human breast cancer. Cancer Res. 66, 1517–1525 (2006).
Robinson, D. R. et al. Functionally recurrent rearrangements of the MAST kinase and Notch gene families in breast cancer. Nature Med. 17, 1646–1651 (2011).
Qiu, M. et al. Specific inhibition of Notch1 signaling enhances the antitumor efficacy of chemotherapy in triple negative breast cancer through reduction of cancer stem cells. Cancer Lett. 328, 261–270 (2013).
Gallahan, D. et al. Expression of a truncated Int3 gene in developing secretory mammary epithelium specifically retards lobular differentiation resulting in tumorigenesis. Cancer Res. 56, 1775–1785 (1996).
Robbins, J., Blondel, B. J., Gallahan, D. & Callahan, R. Mouse mammary tumor gene int-3: a member of the notch gene family transforms mammary epithelial cells. J. Virol. 66, 2594–2599 (1992).
Landor, S. K.-J. et al. Hypo- and hyperactivated Notch signaling induce a glycolytic switch through distinct mechanisms. Proc. Natl Acad. Sci. USA 108, 18814–18819 (2011).
Šale, S., Lafkas, D. & Artavanis-Tsakonas, S. Notch2 genetic fate mapping reveals two previously unrecognized mammary epithelial lineages. Nature Cell Biol. 15, 451–460 (2013).
Parr, C., Watkins, G. & Jiang, W. G. The possible correlation of Notch-1 and Notch-2 with clinical outcome and tumour clinicopathological parameters in human breast cancer. Int. J. Mol. Med. 14, 779–786 (2004).
O'Neill, C. F. et al. Notch2 signaling induces apoptosis and inhibits human MDA-MB-231 xenograft growth. Am. J. Pathol. 171, 1023–1036 (2007).
Stephens, P. J. et al. Whole exome sequencing of adenoid cystic carcinoma. J. Clin. Invest. 123, 2965–2968 (2013).
Deangelo, D. J. et al. A phase I clinical trial of the notch inhibitor MK-0752 in patients with T-cell acute lymphoblastic leukemia/lymphoma (T-ALL) and other leukemias. J. Clin. Oncol. Abstr. 24, 6585 (2006).
Ridgway, J. et al. Inhibition of Dll4 signalling inhibits tumour growth by deregulating angiogenesis. Nature 444, 1083–1087 (2006). This paper, along with reference 241, demonstrates that specific inhibition of DLL4 signalling alters tumour neovascularization such that tumour growth is inhibited in animal models.
Li, J.-L. et al. Delta-like 4 Notch ligand regulates tumor angiogenesis, improves tumor vascular function, and promotes tumor growth in vivo. Cancer Res. 67, 11244–11253 (2007).
Kuramoto, T. et al. Dll4-Fc, an inhibitor of Dll4-Notch signaling, suppresses liver metastasis of small cell lung cancer cells through the downregulation of the NF-κB activity. Mol. Cancer Ther. 11, 2578–2587 (2012).
Jenkins, D. W. et al. MEDI0639: a novel therapeutic antibody targeting Dll4 modulates endothelial cell function and angiogenesis in vivo. Mol. Cancer Ther. 11, 1650–1660 (2012).
Scehnet, J. S. et al. Inhibition of Dll4-mediated signaling induces proliferation of immature vessels and results in poor tissue perfusion. Blood 109, 4753–4760 (2007).
Noguera-Troise, I. et al. Blockade of Dll4 inhibits tumour growth by promoting non-productive angiogenesis. Nature 444, 1032–1037 (2006). This paper, along with reference 236, demonstrates that specific inhibition of DLL4 signalling alters tumour neovascularization such that tumour growth is inhibited in animal models.
Zhao, X.-C. et al. Inhibition of tumor angiogenesis and tumor growth by the DSL domain of human Delta-like 1 targeted to vascular endothelial cells. Neoplasia 15, 815–825 (2013).
Li, K. et al. Modulation of Notch signaling by antibodies specific for the extracellular negative regulatory region of NOTCH3. J. Biol. Chem. 283, 8046–8054 (2008).
Tolcher, Anthony, W. et al. A first-in-human phase I study to evaluate the fully human monoclonal antibody OMP-59R5 (anti-Notch2/3) administered intravenously to patients with advanced solid tumors. J. Clin. Oncol. Abstr. 30, 3025 (2012).
Davis, S. L. et al. Abstract B48: a first-in-human Phase I study of the novel cancer stem cell (CSC) targeting antibody OMP-52M51 (anti-Notch1) administered intravenously to patients with certain advanced solid tumors. Mol. Cancer Ther. 12, B48 (2013).
Doody, R. S. et al. A phase 3 trial of semagacestat for treatment of Alzheimer's disease. N. Engl. J. Med. 369, 341–350 (2013). This study reports on the effects of a GSI (semagacestat) in a Phase III clinical trial for Alzheimer's disease. The trial was terminated before completion, as patients showed off-target effects such as increased incidence of skin cancer and reduced numbers of lymphocytes — effects that are possibly due to perturbed Notch signalling.
Tolcher, A. W. et al. Phase I study of RO4929097, a gamma secretase inhibitor of Notch signaling, in patients with refractory metastatic or locally advanced solid tumors. J. Clin. Oncol. 30, 2348–2353 (2012).
Rosen, L. B. et al. The gamma secretase inhibitor MK-0752 acutely and significantly reduces CSF Abeta40 concentrations in humans. Alzheimers Dement. 2, S79 (2006).
Krop, I. et al. Phase I pharmacologic and pharmacodynamic study of the gamma secretase (Notch) inhibitor MK-0752 in adult patients with advanced solid tumors. J. Clin. Oncol. 30, 2307–2313 (2012).
Kolb, E. A. et al. Initial testing (stage 1) by the pediatric preclinical testing program of RO4929097, a γ-secretase inhibitor targeting notch signaling. Pediatr. Blood Cancer 58, 815–818 (2012).
Luistro, L. et al. Preclinical profile of a potent γ-secretase inhibitor targeting Notch signaling with in vivo efficacy and pharmacodynamic properties. Cancer Res. 69, 7672–7680 (2009).
Strosberg, J. R. et al. A phase II study of RO4929097 in metastatic colorectal cancer. Eur. J. Cancer 48, 997–1003 (2012).
Wang, Z. et al. Targeting Notch signaling pathway to overcome drug resistance for cancer therapy. Biochim. Biophys. Acta 1806, 258–267 (2010).
Yu, S.-D., Liu, F.-Y. & Wang, Q.-R. Notch inhibitor: a promising carcinoma radiosensitizer. Asian Pac. J. Cancer Prev. 13, 5345–5351 (2012).
Timme, C. R., Gruidl, M. & Yeatman, T. J. Gamma-secretase inhibition attenuates oxaliplatin-induced apoptosis through increased Mcl-1 and/or Bcl-xL in human colon cancer cells. Apoptosis 18, 1163–1174 (2013).
Meng, R. D. et al. γ-secretase inhibitors abrogate oxaliplatin-induced activation of the Notch-1 signaling pathway in colon cancer cells resulting in enhanced chemosensitivity. Cancer Res. 69, 573–582 (2009).
Liu, S., Breit, S., Danckwardt, S., Muckenthaler, M. U. & Kulozik, A. E. Downregulation of Notch signaling by γ-secretase inhibition can abrogate chemotherapy-induced apoptosis in T-ALL cell lines. Ann. Hematol. 88, 613–621 (2009).
Sahebjam, S. et al. A phase I study of the combination of RO4929097 and cediranib in patients with advanced solid tumours (PJC-004/NCI 8503). Br. J. Cancer 109, 943–949 (2013).
Diaz-Padilla, I. et al. A phase Ib combination study of RO4929097, a gamma-secretase inhibitor, and temsirolimus in patients with advanced solid tumors. Invest. New Drugs 31, 1182–1191 (2013).
Richter, S. et al. A phase I study of the oral gamma secretase inhibitor R04929097 in combination with gemcitabine in patients with advanced solid tumors (PHL-078/CTEP 8575). Invest. New Drugs 32, 243–249 (2014).
Mizutari, K. et al. Notch inhibition induces cochlear hair cell regeneration and recovery of hearing after acoustic trauma. Neuron 77, 58–69 (2013). This study describes the effects of using a GSI in a mouse model of acoustic trauma to improve hair cell regrowth and induce partial recovery of hearing.
Slowik, A. D. & Bermingham-McDonogh, O. Hair cell generation by Notch inhibition in the adult mammalian cristae. J. Assoc. Res. Otolaryngol. 14, 813–828 (2013).
Jin, S. et al. Notch signaling regulates platelet-derived growth factor receptor-β expression in vascular smooth muscle cells. Circ. Res. 102, 1483–1491 (2008).
Eickelberg, O. & Morty, R. E. Transforming growth factor β/bone morphogenic protein signaling in pulmonary arterial hypertension: remodeling revisited. Trends Cardiovasc. Med. 17, 263–269 (2007).
O'Callaghan, D. S. et al. Treatment of pulmonary arterial hypertension with targeted therapies. Nature Rev. Cardiol. 8, 526–538 (2011).
Tran, I. T. et al. Blockade of individual Notch ligands and receptors controls graft-versus-host disease. J. Clin. Invest. 123, 1590–1604 (2013). This report demonstrates that blocking DLL1 and DLL4 reduces GVHD without having adverse side effects in mouse models. A transient blockade was shown to be sufficient for durable protection.
Chen, Y. et al. Inhibition of Notch signaling by a γ-secretase inhibitor attenuates hepatic fibrosis in rats. PLoS ONE 7, e46512 (2012).
Siemers, E. R. et al. Effects of a γ-secretase inhibitor in a randomized study of patients with Alzheimer disease. Neurology 66, 602–604 (2006).
Fleisher, A. S. et al. Phase 2 safety trial targeting amyloid β production with a γ-secretase inhibitor in Alzheimer disease. Arch. Neurol. 65, 1031–1038 (2008).
Pandya, K. et al. Targeting both Notch and ErbB-2 signalling pathways is required for prevention of ErbB-2-positive breast tumour recurrence. Br. J. Cancer 105, 796–806 (2011).
Farnie, G., Willan, P. M., Clarke, R. B. & Bundred, N. J. Combined inhibition of ErbB1/2 and Notch receptors effectively targets breast ductal carcinoma in situ (DCIS) stem/progenitor cell activity regardless of ErbB2 status. PLoS ONE 8, e56840 (2013).
Ma, Y. et al. Inhibition of the Wnt-β-catenin and Notch signaling pathways sensitizes osteosarcoma cells to chemotherapy. Biochem. Biophys. Res. Commun. 431, 274–279 (2013).
Jin, R. et al. Combination therapy using Notch and Akt inhibitors is effective for suppressing invasion but not proliferation in glioma cells. Neurosci. Lett. 534, 316–321 (2013).
Shepherd, C. et al. PI3K/mTOR inhibition upregulates NOTCH–MYC signalling leading to an impaired cytotoxic response. Leukemia 27, 650–660 (2013).
Yao, J. et al. Combination treatment of PD98059 and DAPT in gastric cancer through induction of apoptosis and downregulation of WNT/β-catenin. Cancer Biol. Ther. 14, 833–839 (2013).
Slowing, I. I., Trewyn, B. G., Giri, S. & Lin, V. S. Y. Mesoporous silica nanoparticles for drug delivery and biosensing applications. Adv. Funct. Mater. 17, 1225–1236 (2007).
Mamaeva, V. et al. Mesoporous silica nanoparticles as drug delivery systems for targeted inhibition of Notch signaling in cancer. Mol. Ther. J. Am. Soc. Gene Ther. 19, 1538–1546 (2011).
Beckstead, B. L. et al. Methods to promote Notch signaling at the biomaterial interface and evaluation in a rafted organ culture model. J. Biomed. Mater. Res. Part A 91, 436–446 (2009).
Kovall, R. A. & Blacklow, S. C. Mechanistic insights into Notch receptor signaling from structural and biochemical studies. Curr. Top. Dev. Biol. 92, 31–71 (2010).
Hock, B. et al. PDZ-domain-mediated interaction of the Eph-related receptor tyrosine kinase EphB3 and the ras-binding protein AF6 depends on the kinase activity of the receptor. Proc. Natl Acad. Sci. USA 95, 9779–9784 (1998).
LaVoie, M. J. & Selkoe, D. J. The Notch ligands, Jagged and Delta, are sequentially processed by α-secretase and presenilin/γ-secretase and release signaling fragments. J. Biol. Chem. 278, 34427–34437 (2003).
Higaki, J., Quon, D., Zhong, Z. & Cordell, B. Inhibition of β-amyloid formation identifies proteolytic precursors and subcellular site of catabolism. Neuron 14, 651–659 (1995).
Barthet, G., Georgakopoulos, A. & Robakis, N. K. Cellular mechanisms of γ-secretase substrate selection, processing and toxicity. Prog. Neurobiol. 98, 166–175 (2012).
Wolfe, M. S. γ-secretase as a target for Alzheimer's disease. Adv. Pharmacol. 64, 127–153 (2012).
Louvi, A. & Artavanis-Tsakonas, S. Notch and disease: a growing field. Semin. Cell Dev. Biol. 23, 473–480 (2012).
Willander, K. et al. NOTCH1 mutations influence survival in chronic lymphocytic leukemia patients. BMC Cancer 13, 274 (2013).
Zong, Y. & Stanger, B. Z. Molecular mechanisms of bile duct development. Int. J. Biochem. Cell Biol. 43, 257–264 (2011).
Massi, D. & Panelos, J. Notch signaling and the developing skin epidermis. Adv. Exp. Med. Biol. 727, 131–141 (2012).
Martignetti, J. A. et al. Mutations in PDGFRB cause autosomal-dominant infantile myofibromatosis. Am. J. Hum. Genet. 92, 1001–1007 (2013).
Lee, J. Mutations in PDGFRB and NOTCH3 are the first genetic causes identified for autosomal dominant infantile myofibromatosis. Clin. Genet. 84, 340–341 (2013).
Michailidis, C. et al. Notch2, Notch4 gene polymorphisms in psoriasis vulgaris. Eur. J. Dermatol. 23, 146–153 (2013).
Sun, W. et al. Activation of the NOTCH pathway in head and neck cancer. Cancer Res. 74, 1091–1104 (2013).
Wang, N. J. et al. Loss-of-function mutations in Notch receptors in cutaneous and lung squamous cell carcinoma. Proc. Natl Acad. Sci. USA 108, 17761–17766 (2011).
Agrawal, N. et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science 333, 1154–1157 (2011).
Stransky, N. et al. The mutational landscape of head and neck squamous cell carcinoma. Science 333, 1157–1160 (2011).
Dunwoodie, S. L. The role of Notch in patterning the human vertebral column. Curr. Opin. Genet. Dev. 19, 329–337 (2009).
Regan, J. & Long, F. Notch signaling and bone remodeling. Curr. Osteoporos. Rep. 11, 126–129 (2013).
Fouillade, C., Monet-Leprêtre, M., Baron-Menguy, C. & Joutel, A. Notch signalling in smooth muscle cells during development and disease. Cardiovasc. Res. 95, 138–146 (2012).
Benedito, R. & Hellström, M. Notch as a hub for signaling in angiogenesis. Exp. Cell Res. 319, 1281–1288 (2013).
Acknowledgements
E.R.A. holds a junior research grant from the Swedish Research Council. Work in U.L.'s laboratory is funded by the Swedish Research Council (DBRM, StratRegen and project grant), the Swedish Cancer Society, Knut och Alice Wallenbergs Stiftelse and Karolinska Institutet (the Breast Cancer Theme Center (BRECT) and a Distinguished Professor Award).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information S1 (table)
Ongoing and completed studies with Notch targeting (from clinicaltrials.gov), same as Table 3, but including stated purpose of trial. (PDF 231 kb)
Glossary
- Signal-sending cell
-
The cell on which the ligand is expressed. Canonical Notch signalling is seen as a uni-directional signalling pathway in which the ligand is expressed on one cell, which 'sends' the signal, and the receptor is expressed on a contacting cell, which 'receives' the signal.
- Signal-receiving cell
-
The cell on which the receptor is expressed. Canonical Notch signalling is seen as a uni-directional signalling pathway in which the ligand is expressed on one cell, which 'sends' the signal, and the receptor is expressed on a contacting cell, which 'receives' the signal.
- Epithelial to mesenchymal transition
-
(EMT). A phenotypic change in cells where they lose epithelial and/or adhesive properties and become more motile. This is often associated with the onset of cancer metastasis.
- Wolff–Parkinson–White syndrome
-
A heart condition in which aberrant conductance between atria and ventricles leads to an increased heart rate.
- Maximum tolerated dose
-
The highest dose of a treatment that will produce the desired effect without causing unacceptable toxicity.
Rights and permissions
About this article
Cite this article
Andersson, E., Lendahl, U. Therapeutic modulation of Notch signalling — are we there yet?. Nat Rev Drug Discov 13, 357–378 (2014). https://doi.org/10.1038/nrd4252
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrd4252
This article is cited by
-
Notch activator cyclopiazonic acid induces apoptosis in HL-60 cells through calcineurin activation
The Journal of Antibiotics (2024)
-
GRWD1 Over-Expression Promotes Gastric Cancer Progression by Activating Notch Signaling Pathway via Up-Regulation of ADAM17
Digestive Diseases and Sciences (2024)
-
Cross-talk between cancer stem cells and immune cells: potential therapeutic targets in the tumor immune microenvironment
Molecular Cancer (2023)
-
Antibody blockade of Jagged1 attenuates choroidal neovascularization
Nature Communications (2023)
-
Extensive review on breast cancer its etiology, progression, prognostic markers, and treatment
Medical Oncology (2023)