Key Points
-
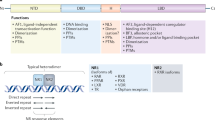
The nuclear receptors retinoic acid receptor-related orphan receptor-α (RORα), RORβ, RORγ, REV-ERBα and REV-ERBβ were originally identified as orphan receptors. RORα and RORβ constitutively activate transcription, whereas REV-ERBα and REV-ERBβ constitutively silence transcription.
-
RORα and RORγ are now known to bind to sterols, with certain oxysterols having a very high affinity for these receptors. REV-ERBs have been found to bind to haem.
-
REV-ERBs function as ligand-dependent (that is, haem-dependent) silencers of transcription. The role of the endogenous ligands for the RORs is less clear, as several sterols and oxysterols have been suggested to function as agonists or inverse agonists.
-
The RORs and REV-ERBs have substantially overlapping functions as they usually recognize similar DNA response elements. These receptors have important roles in many physiological functions, including development, circadian rhythm, metabolism and immune function.
-
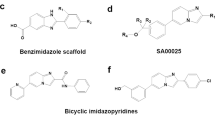
Over the past several years, synthetic ligands have been designed that target RORs and REV-ERBs. Many of these have high potency and have been used to examine the utility of targeting RORs and REV-ERBs in animal models of human disease.
-
Synthetic REV-ERB agonists alter the circadian rhythm and have beneficial effects on the metabolic profile in obese mice. REV-ERB agonists increase oxidative metabolism in the skeletal muscle and improve exercise endurance in mice.
-
Synthetic inverse agonists of ROR (that either target RORγ alone or both RORα and RORγ) are effective in treating and preventing autoimmunity in mouse models. Additionally, they have beneficial effects on glucose and lipid metabolism.
-
The continued refinement and development of synthetic ligands that target these former orphan nuclear receptors may yield novel therapeutics to treat a range of diseases in the future.
Abstract
The nuclear receptors REV-ERB (consisting of REV-ERBα and REV-ERBβ) and retinoic acid receptor-related orphan receptors (RORs; consisting of RORα, RORβ and RORγ) are involved in many physiological processes, including regulation of metabolism, development and immunity as well as the circadian rhythm. The recent characterization of endogenous ligands for these former orphan nuclear receptors has stimulated the development of synthetic ligands and opened up the possibility of targeting these receptors to treat several diseases, including diabetes, atherosclerosis, autoimmunity and cancer. This Review focuses on the latest developments in ROR and REV-ERB pharmacology indicating that these nuclear receptors are druggable targets and that ligands targeting these receptors may be useful in the treatment of several disorders.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Mangelsdorf, D. J. et al. The nuclear receptor superfamily — the 2nd decade. Cell 83, 835–839 (2005).
McKenna, N. J., Lanz, R. B. & O'Malley, B. W. Nuclear receptor coregulators: cellular and molecular biology. Endocr. Rev. 20, 321–344 (1999).
Savkur, R. S. & Burris, T. P. The coactivator LXXLL nuclear receptor recognition motif. J. Pept. Res. 63, 207–212 (2004).
Kliewer, S. A., Lehmann, J. M. & Willson, T. M. Orphan nuclear receptors: shifting endocrinology into reverse. Science 284, 757–760 (1999).
Schulman, I. G. & Heyman, R. A. The flip side: Identifying small molecule regulators of nuclear receptors. Chem. Biol. 11, 639–646 (2004).
Burris, T. P., Busby, S. A. & Griffin, P. R. Targeting orphan nuclear receptors for treatment of metabolic diseases and autoimmunity. Chem. Biol. 19, 51–59 (2012).
Burris, T. P. Nuclear hormone receptors for heme: REV-ERBα and REV-ERBβ are ligand-regulated components of the mammalian clock. Mol. Endocrinol. 22, 1509–1520 (2008).
Jetten, A. M. Retinoid-related orphan receptors (RORs): critical roles in development, immunity, circadian rhythm, and cellular metabolism. Nucl. Recept. Signal. 7, e003 (2009).
Jetten, A. M., Kang, H. S. & Takeda, Y. Retinoic acid-related orphan receptors α and γ: key regulators of lipid/glucose metabolism, inflammation, and insulin sensitivity. Frontiers Endocrinol. 4, 1 (2013).
Duez, H. & Staels, B. Rev-erb-α: an integrator of circadian rhythms and metabolism. J. Appl. Physiol. 107, 1972–1980 (2009).
Lazar, M. A., Hodin, R. A., Darling, D. S. & Chin, W. W. A novel member of the thyroid steroid hormone receptor family is encoded by the opposite strand of the rat c-erbA-α transcriptional unit. Mol. Cell. Biol. 9, 1128–1136 (1989).
Miyajima, N. et al. 2 Erba homologs encoding proteins with different T3 binding-capacities are transcribed from opposite DNA strands of the same genetic-locus. Cell 57, 31–39 (1989).
Miyajima, N. et al. Identification of 2 novel members of Erba superfamily by molecular-cloning - the gene-products of the 2 are highly related to each other. Nucleic Acids Res. 16, 11057–11074 (1988).
Dumas, B. et al. A new orphan member of the nuclear hormone receptor superfamily closely related to REV-ERB. Mol. Endocrinol. 8, 996–1005 (1994).
Forman, B. M. et al. Cross talk among RORα1 and the REV-ERB family of orphan nuclear receptors. Mol. Endocrinol. 8, 1253–1261 (1994).
Bonnelye, E. et al. Rev-erb-β, a new member of the nuclear receptor superfamily, is expressed in the nervous-system during chicken development. Cell Growth Differ. 5, 1357–1365 (1994).
Yin, L. & Lazar, M. A. The orphan nuclear receptor Rev-erbα recruits the N-CoR/histone deacetylase 3 corepressor to regulate the circadian Bmal1 gene. Mol. Endocrinol. 19, 1452–1459 (2005).
Harding, H. P. & Lazar, M. A. The monomer-binding orphan receptor rev-erb represses transcription as a dimer on a novel direct repeat. Mol. Cell. Biol. 15, 6479–6479 (1995).
Harding, H. P. & Lazar, M. A. The orphan receptor rev-erba-α activates transcription via a novel response element. Mol. Cell. Biol. 13, 3113–3121 (1993).
Zvonic, S. et al. Characterization of peripheral circadian clocks in adipose tissues. Diabetes 55, 962–970 (2006).
Balsalobre, A., Damiola, F. & Schibler, U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell 93, 929–937 (1998).
Guillaumond, F., Dardente, H., Giguere, V. & Cermakian, N. Differential control of Bmal1 circadian transcription by REV-ERB and ROR nuclear receptors. J. Biol. Rhythms 20, 391–403 (2005).
Torra, I. P. et al. Circadian and glucocorticoid regulation of Rev-erbα expression in liver. Endocrinology 141, 3799–3806 (2000).
Beckerandre, M., Andre, E. & Delamarter, J. F. Identification of nuclear receptor messenger RNAs by RT-PCR amplification of conserved zinc finger motif sequences. Biochem. Biophys. Res. Commun. 194, 1371–1379 (1993).
Giguere, V. et al. Isoform specific amino-terminal domains dictate DNA binding properties of RORα, a novel family of orphan nuclear receptors. Genes Dev. 8, 538–553 (1994).
Carlberg, C., Vanhuijsduijnen, R. H., Staple, J. K., Delamarter, J. F. & Beckerandre, M. RZRs, a new family of retinoid-related orphan receptors that function as both monomers heterodimers. Mol. Endocrinol. 8, 757–770 (1994).
Hirose, T., Smith, R. J. & Jetten, A. M. RORγ - the 3rd member of ROR-RZR orphan receptor subfamily that is highly expressed in skeletal muscle. Biochem. Biophys. Res. Commun. 205, 1976–1983 (1994).
Hamilton, B. A. et al. Disruption of the nuclear hormone receptor RORα in staggerer mice. Nature 379, 736–739 (1996).
Steinmayr, M. et al. staggerer phenotype in retinoid-related orphan receptor α-deficient mice. Proc. Natl Acad. Sci. USA 95, 3960–3965 (1998).
Andre, E., Gawlas, K., Steinmayr, M. & Becker-Andre, M. A novel isoform of the orphan nuclear receptor ROR β is specifically expressed in pineal gland and retina. Gene 216, 277–283 (1998).
Smith, A. G. & Muscat, G. E. O. Orphan nuclear receptors: therapeutic opportunities in skeletal muscle. Am. J. Physiol. Cell Physiol. 291, C203–C217 (2006).
Mohawk, J. A., Green, C. B. & Takahashi, J. S. Central and peripheral circadian clocks in mammals. Annu. Rev. Neurosci. 35, 445–462 (2012).
Yang, X. Y. et al. Cell 126, 801–810 (2006).
Lamia, K. A. et al. Cryptochromes mediate rhythmic repression of the glucocorticoid receptor. Nature 480, 552–556 (2011).
Schmutz, I., Ripperger, J. A., Baeriswyl-Aebischer, S. & Albrecht, U. The mammalian clock component PERIOD2 coordinates circadian output by interaction with nuclear receptors. Genes Dev. 24, 345–357 (2010).
Preitner, N. et al. The orphan nuclear receptor REV-ERBα controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell 110, 251–260 (2002). This paper describes the first characterization of a role for REV-ERB in the regulation of the circadian rhythm.
Triqueneaux, G. et al. The orphan receptor Rev-erbα gene is a target of the circadian clock pacemaker. J. Mol. Endocrinol. 33, 585–608 (2004).
Ripperger, J. A. Mapping of binding regions for the circadian regulators BMAL1 and CLOCK within the mouse Rev-erbα gene. Chronobiol. Int. 23, 135–142 (2006).
Cho, H. et al. Regulation of circadian behaviour and metabolism by REV-ERB-α and REV-ERB-β. Nature 485, 123–127 (2012). This study defines the critical overlapping role of REV-ERBα and REV-ERBβ in the control of the circadian rhythm.
Bunger, M. K. et al. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell 103, 1009–1017 (2000).
Vitaterna, M. H. et al. Differential regulation of mammalian period genes and circadian rhythmicity by cryptochromes 1 and 2. Proc. Natl Acad. Sci. USA 96, 12114–12119 (1999).
Bae, K. et al. Differential functions of mPer1, mPer2, and mPer3 in the SCN circadian clock. Neuron 30, 525–536 (2001).
Sato, T. K. et al. A functional genomics strategy reveals rora as a component of the mammalian circadian clock. Neuron 43, 527–537 (2004).
Andre, E. et al. Disruption of retinoid-related orphan receptor β changes circadian behavior, causes retinal degeneration and leads to vacillans phenotype in mice. EMBO J. 17, 3867–3877 (1998).
Masana, M. I., Sumaya, I. C., Becker-Andre, M. & Dubocovich, M. L. Behavioral characterization and modulation of circadian rhythms by light and melatonin in C3H/HeN mice homozygous for the RORβ knockout. Am. J. Physiol. Regul. Integr. Comp. Physiol. 292, R2357–R2367 (2007).
Takeda, Y., Jothi, R., Birault, V. & Jetten, A. M. RORγ directly regulates the circadian expression of clock genes and downstream targets in vivo. Nucleic Acids Res. 40, 8519–8535 (2012).
Bass, J. & Takahashi, J. S. Circadian integration of metabolism and energetics. Science 330, 1349–1354 (2010).
Green, C. B., Takahashi, J. S. & Bass, J. The meter of metabolism. Cell 134, 728–742 (2008).
Gamble, K. L. & Young, M. E. Metabolism as an integral cog in the mammalian circadian clockwork. Crit. Rev. Biochem. Mol. Biol. (2013).
Eckel-Mahan, K. & Sassone-Corsi, P. Metabolism and the circadian clock converge. Physiol. Rev. 93, 107–135 (2013).
Bass, J. Circadian topology of metabolism. Nature 491, 348–356 (2012).
Li, Y., Sato, Y. & Yamaguchi, N. Shift work and the risk of metabolic syndrome: a nested case-control study. Int. J. Occupat. Environ. Health 17, 154–160 (2011).
Baron, K. G., Reid, K. J., Kern, A. S. & Zee, P. C. Role of sleep timing in caloric intake and BMI. Obesity 19, 1374–1381 (2011).
Karlsson, B., Knutsson, A. & Lindahl, B. Is there an association between shift work and having a metabolic syndrome? Results from a population based study of 27,485 people. Occupat. Environ. Med. 58, 747–752 (2001).
Scheer, F. A. J. L., Hilton, M. F., Mantzoros, C. S. & Shea, S. A. Adverse metabolic and cardiovascular consequences of circadian misalignment. Proc. Natl Acad. Sci. USA 106, 4453–4458 (2009).
Markwald, R. R. et al. Impact of insufficient sleep on total daily energy expenditure, food intake, and weight gain. Proc. Natl Acad. Sci. USA 110, 5695–5700 (2013).
Raspe, E. et al. Identification of Rev-erbα as a physiological repressor of apoC-III gene transcription. J. Lipid Res. 43, 2172–2179 (2002).
Raspe, E., Mautino, G., Duez, H., Fruchart, J. C. & Staels, B. Transcriptional regulation of apolipoprotein C-III gene expression by the orphan nuclear receptor Rev-erbα. Circulation 104, 15–15 (2001).
Vu-Dac, N. et al. The nuclear receptors peroxisome proliferator-activated receptorα and Rev-erbα mediate the species-specific regulation of apolipoprotein A-I expression by fibrates. J. Biol. Chem. 273, 25713–25720 (1998).
Ramakrishnan, S. N., Lau, P., Burke, L. J. & Muscat, G. E. O. Rev-erbβ regulates the expression of genes involved in lipid absorption in skeletal muscle cells — evidence for cross-talk between orphan nuclear receptors and myokines. J. Biol. Chem. 280, 8651–8659 (2005).
Yin, L. et al. Rev-erbα, a heme sensor that coordinates metabolic and circadian pathways. Science 318, 1786–1789 (2007). Along with reference 84, this study defines haem as an endogenous ligand of REV-ERB.
Delezie, J. et al. The nuclear receptor REV-ERBα is required for the daily balance of carbohydrate and lipid metabolism. FASEB J. 26, 3321–3335 (2012).
Kang, H. S. et al. Gene expression profiling reveals a regulatory role for RORα and RORγ in phase I and phase II metabolism. Physiol. Genom. 31, 281–294 (2007).
Lau, P., Fitzsimmons, R. L., Pearen, M. A., Watt, M. J. & Muscat, G. E. Homozygous staggerer (sg/sg) mice display improved insulin sensitivity and enhanced glucose uptake in skeletal muscle. Diabetologia 54, 1169–1180 (2011).
Woldt, E. et al. Rev-erba modulates skeletal muscle oxidative capacity by regulated mitochondrial biogenesis and autophagy. Nature Med. 19, 1039–1046 (2013). This paper demonstrates that REV-ERB has a role in the regulation of the oxidative capacity of skeletal muscle, and also shows that SR9009 treatment could increase exercise endurance in mice.
Lau, P., Nixon, S. J., Parton, R. G. & Muscat, G. E. O. RORα regulates the expression of genes involved in lipid homeostasis in skeletal muscle cells: caveolin-3 and CPT-1 are direct targets of ROR. J. Biol. Chem. 279, 36828–36840 (2004).
Chawla, A. & Lazar, M. A. Induction of REV-ERBα, an orphan receptor encoded on the opposite strand of the α thyroid hormone receptor gene, during adipocyte differentiation. J. Biol. Chem. 268, 16265–16269 (1993).
Fontaine, C. et al. The orphan nuclear receptor Rev-Erbα is a peroxisome proliferator-activated receptor (PPAR) γ target gene and promotes PPARγ-induced adipocyte differentiation. J. Biol. Chem. 278, 37672–37680 (2003).
Wang, J. & Lazar, M. A. Bifunctional role of Rev-erbα in adipocyte differentiation. Mol Cell. Biol 28, 2213–2220 (2008).
Kojetin, D. J. & Burris, T. P. A role for rev-erbα ligands in regulation of adipogenesis. Curr. Pharm. Design 17, 320–324 (2011).
Kumar, N. et al. Regulation of adipogenesis by natural and synthetic REV-ERB ligands. Endocrinology 151, 3015–3025 (2010).
Lau, P. et al. The orphan nuclear receptor, RORα, regulates gene expression that controls lipid metabolism: staggerer (sg/sg) mice are resistant to diet-induced obesity. J. Biol. Chem. 283, 18411–18421 (2008).
Meissburger, B. et al. Adipogenesis and insulin sensitivity in obesity are regulated by retinoid-related orphan receptor γ. EMBO Mol. Med. 3, 637–651 (2011).
Feng, D. et al. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science 331, 1315–1319 (2011).
Bettelli, E., Oukka, M. & Kuchroo, V. K. TH-17 cells in the circle of immunity and autoimmunity. Nature Immunol. 8, 345–350 (2007).
Fouser, L. A., Wright, J. F., Dunussi-Joannopoulos, K. & Collins, M. Th17 cytokines and their emerging roles in inflammation and autoimmunity. Immunol. Rev. 226, 87–102 (2008).
Ivanov, I. I. et al. The orphan nuclear receptor RORγt directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell 126, 1121–1133 (2006). This study describes the crucial role of RORγ in the differentiation of T H 17 cells.
Yang, X. X. O. et al. T helper 17 lineage differentiation is programmed by orphan nuclear receptors RORα and RORγ. Immunity 28, 29–39 (2008).
Gibbs, J. E. et al. The nuclear receptor REV-ERBα mediates circadian regulation of innate immunity through selective regulation of inflammatory cytokines. Proc. Natl Acad. Sci. USA 109, 582–587 (2012).
Lam, M. T. et al. Rev-Erbs repress macrophage gene expression by inhibiting enhancer-directed transcription. Nature 498, 511–515 (2013).
Yu, X. et al. TH17 cell differentiation is regulated by the circadian clock. Science 342, 727–730 (2013).
Ma, H. et al. Increased atherosclerotic lesions in LDL receptor deficient mice with hematopoietic nuclear receptor Rev-erbα knock- down. J. Am. Heart Assoc. 2, e000235 (2013).
Reinking, J. et al. The Drosophila nuclear receptor E75 contains heme and is gas responsive. Cell 122, 195–207 (2005).
Raghuram, S. et al. Identification of heme as the ligand for the orphan nuclear receptors REV-ERBα and REV-ERBβ. Nature Struct. Mol. Biol. 14, 1207–1213 (2007). Along with reference 61, this study defines haem as an endogenous ligand of REV-ERB.
Wu, N., Yin, L., Hanniman, E. A., Joshi, S. & Lazar, M. A. Negative feedback maintenance of heme homeostasis by its receptor, Rev-erbα. Genes Dev. 23, 2201–2209 (2009).
Estall, J. L. et al. PGC-1α negatively regulates hepatic FGF21 expression by modulating the heme/Rev-Erbα axis. Proc. Natl Acad. Sci. USA 106, 22510–22515 (2009).
Rogers, P. M., Ying, L. & Burris, T. P. Relationship between circadian oscillations of Rev-erbα expression and intracellular levels of its ligand, heme. Biochem. Biophys. Res. Commun. 368, 955–958 (2008).
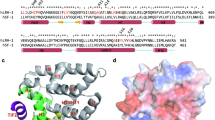
Pardee, K. I. et al. The structural basis of gas-responsive transcription by the human nuclear hormone receptor REV-ERBβ. Plos Biol. 7, 384–398 (2009).
Gupta, N. & Ragsdale, S. W. Thiol-disulfide redox dependence of heme binding and heme ligand switching in nuclear hormone receptor rev-erbβ. J. Biol. Chem. 286, 4392–4403 (2011).
Phelan, C. A. et al. Structure of Rev-erbα bound to N-CoR reveals a unique mechanism of nuclear receptor-co-repressor interaction. Nature Struct. Mol. Biol. 17, 808–814 (2010).
Marvin, K. A. et al. Nuclear receptors Homo sapiens rev-erb beta and Drosophila melanogaster E75 are thiolate-ligated heme proteins which undergo redox-mediated ligand switching and bind CO and NO. Biochemistry 48, 7056–7071 (2009).
Golombek, D. A., Agostino, P. V., Plano, S. A. & Ferreyra, G. A. Signaling in the mammalian circadian clock: the NO/cGMP pathway. Neurochem. Int. 45, 929–936 (2004).
Kallen, J. A. et al. X-ray structure of the hRORα LBD at 1.63 Å: structural and functional data that cholesterol or a cholesterol derivative is the natural ligand of RORα. Structure 10, 1697–1707 (2002). This study uses X-ray crystallography to show that sterols could bind to RORα.
Kallen, J., Schlaeppi, J. M., Bitsch, F., Delhon, I. & Fournier, B. Crystal structure of the human RORα ligand binding domain in complex with cholesterol sulfate at 2.2 angstrom. J. Biol. Chem. 279, 14033–14038 (2004).
Wang, Y., Kumar, N., Crumbley, C., Griffin, P. R. & Burris, T. P. A second class of nuclear receptors for oxysterols: regulation of RORα and RORγ activity by 24S-hydroxycholesterol (cerebrosterol). Biochim. Biophys. Acta 1801, 917–923 (2010).
Wang, Y. et al. Modulation of RORα and RORγ activity by 7-oxygenated sterol ligands. J. Biol. Chem. 285, 5013–5025 (2010). This paper defines a role for oxysterols as ligands for RORα and RORγ.
Jin, L. H. et al. Structural basis for hydroxycholesterols as natural ligands of orphan nuclear receptor RORγ. Mol. Endocrinol. 24, 923–929 (2010). This paper identifies a role for oxysterols as ligands for RORγ.
Stehlin-Gaon, C. et al. All-trans retinoic acid is a ligand for the orphan nuclear receptor RORβ. Nature Struct. Biol. 10, 820–825 (2003).
Helleboid, S. et al. The identification of naturally occurring neoruscogenin as a bioavailable, potent, and high-affinity agonist of the nuclear receptor RORα (NR1F1). J. Biomol. Screen. http://dx.doi.org/10.1177/1087057113497095 (2013).
Stehlin, C. et al. X-ray structure of the orphan nuclear receptor RORβ ligand-binding domain in the active conformation. EMBO J. 20, 5822–5831 (2001).
Meng, Q. J. et al. Ligand modulation of REV-ERBα function resets the peripheral circadian clock in a phasic manner. J. Cell Sci. 121, 3629–3635 (2008). This was the first published description of a synthetic REV-ERB agonist.
Grant, D. et al. GSK4112, a small molecule chemical probe for the cell biology of the nuclear heme receptor Rev-erbα. ACS Chem. Biol. 5, 925–932 (2010). This is a description of the discovery of the first synthetic REV-ERB ligand.
Noel, R. et al. Synthesis and SAR of tetrahydroisoquinolines as Rev-erbα agonists. Bioorg. Med. Chem. Lett. 22, 3739–3742 (2012).
Shin, Y. et al. Small molecule tertiary amines as agonists of the nuclear hormone receptor Rev-erbα. Bioorg. Med. Chem. Lett. 22, 4413–4417 (2012).
Solt, L. A. et al. Regulation of circadian behavior and metabolism by synthetic REV-ERB agonists. Nature 485, 62–68 (2012). This paper describes the discovery of SR9009 and SR9011, the first REV-ERB agonsts, which were used to define the effects of REV-ERB agonists on the circadian rhythm and metabolism in mice.
Yoo, S. H. et al. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc. Natl Acad. Sci. USA 101, 5339–5346 (2004).
Trump, R. P. et al. Optimized chemical probes for REV-ERBα. J. Med. Chem. 56, 4729–4737 (2013).
Kojetin, D., Wang, Y., Kamenecka, T. M. & Burris, T. P. Identification of SR8278, a synthetic antagonist of the nuclear heme receptor, REV-ERB. ACS Chem. Biol. 6, 131–134 (2011). This study describes the first synthetic REV-ERB antagonist.
Vieira, E. et al. The clock gene Rev-erbα regulates pancreatic β-cell function: modulation by leptin and high-fat diet. Endocrinology 153, 592–601 (2012).
Negoro, H. et al. Role of Rev-erbα domains for transactivation of the connexin43 promoter with Sp1. FEBS Lett. 587, 98–103 (2013).
Wiesenberg, I. et al. Specific activation of the nuclear receptors PPARγ and RORA by the antidiabetic thiazolidinedione BRL 49653 and the antiarthritic thiazolidinedione derivative CGP 52608. Mol. Pharmacol. 53, 1131–1138 (1998).
Wiesenberg, I., Missbach, M., Kahlen, J. P., Schrader, M. & Carlberg, C. Transcriptional activation of the nuclear receptor RZRα by the pineal-gland hormone melatonin and identification of cgp-52608 as a synthetic ligand. Nucleic Acids Res. 23, 327–333 (1995).
Missbach, M. et al. Thiazolidine diones, specific ligands of the nuclear receptor retinoid Z receptor/retinoid acid receptor-related orphan receptor α with potent antiarthritic activity. J. Biol. Chem. 271, 13515–13522 (1996).
Park, Y. et al. N-methylthioureas as new agonists of retinoic acid receptor-related orphan receptor. Arch. Pharm. Res. 35, 1393–1401 (2012).
Kumar, N. et al. The benzenesulfonamide T0901317 is a novel RORα/γ inverse agonist. Mol. Pharmacol. 77, 228–236 (2010).
Schultz, J. R. et al. Role of LXRs in control of lipogenesis. Genes Dev. 14, 2831–2838 (2000).
Houck, K. A. et al. T0901317 is a dual LXR/FXR agonist. Mol. Genet. Metab. 83, 184–187 (2004).
Mitro, N., Vargas, L., Romeo, R., Koder, A. & Saez, E. T0901317 is a potent PXR ligand: implications for the biology ascribed to LXR. FEBS Lett. 581, 1721–1726 (2007).
Wang, Y. et al. Identification of SR1078, a synthetic agonist for the orphan nuclear receptors RORA and RORG. ACS Chem. Biol. 5, 1029–1034 (2010).
Wang, Y. J., Solt, L. A. & Burris, T. P. Regulation of FGF21 expression and secretion by retinoic acid receptor-related orphan receptor α. J. Biol. Chem. 285, 15668–15673 (2010).
Kharitonenkov, A. et al. FGF-21 as a novel metabolic regulator. J. Clin. Invest. 115, 1627–1635 (2005).
Kharitonenkov, A. et al. FGF-21 regulates the metabolic state of diabetic monkeys. Diabetes 56, A668–A668 (2007).
Zhu, Y., McAvoy, S., Kuhn, R. & Smith, D. I. RORA, a large common fragile site gene, is involved in cellular stress response. Oncogene 25, 2901–2908 (2006).
Wang, Y., Solt, L. A., Kojetin, D. J. & Burris, T. P. Regulation of p53 stability and apoptosis by a ROR agonist. PloS ONE 7, e34921 (2012).
Kumar, N. et al. Identification of SR3335 (ML-176): a synthetic RORα selective inverse agonist. ACS Chem. Biol. 6, 218–222 (2011).
Solt, L. A. et al. Suppression of TH17 differentiation and autoimmunity by a synthetic ROR ligand. Nature 472, 491–494 (2011).
Xu, J., Wagoner, G., Douglas, J. C. & Drew, P. D. Liver X receptor agonist regulation of Th17 lympocyte function in autoimmunity. J. Leukoc. Biol. 86, 401–409 (2009).
Xu, J. H., Racke, M. K. & Drew, P. D. Peroxisome proliferator-activated receptor-α agonist fenofibrate regulates IL-12 family cytokine expression in the CNS: relevance to multiple sclerosis. J. Neurochem. 103, 1801–1810 (2007).
Kumar, N. et al. Identification of SR2211: a potent synthetic RORγ-selective modulator. ACS Chem. Biol. 7, 672–677 (2012).
Solt, L. A. et al. Identification of a selective RORγ ligand that suppresses TH17 cells and stimulates T regulatory cells. ACS Chem. Biol. 7, 1515–1519 (2012). This paper demonstrates that at least a subgroup of RORγ inverse agonists stimulate the differentiation of T Reg cells as well as suppressing T H 17 cell polarization.
Huh, J. R. et al. Digoxin and its derivatives suppress TH17 cell differentiation by antagonizing RORγt activity. Nature 472, 486–490 (2011). Along with reference 126, this study demonstrates that a RORγ inverse agonist suppresses T H 17 cell development and has potential for the treatment of autoimmune disorders.
Fujita-Sato, S. et al. Structural basis of digoxin that antagonizes RORγt receptor activity and suppresses TH17 cell differentiation and interleukin (IL)-17 production. J. Biol. Chem. 286, 31409–31417 (2011).
Eade, E., Cooper, R. & Mitchell, A. R. Digoxin — time to take the gloves off? Int. J. Cardiol. 164, 365–367 (2013).
Matsui, H. & Schwartz, A. Mechanism of cardiac glycoside inhibition of the (Na+-K+)-dependent ATPase from cardiac tissue. Biochim. Biophys. Acta 151, 655–663 (1968).
Xu, T. et al. Ursolic acid suppresses IL-17 production by selectively antagonizing the function of RORγt protein. J. Biol. Chem. 286, 22707–22710 (2011).
He, Y., Li, Y., Zhao, T., Wang, Y. & Sun, C. Ursolic acid inhibits adipogenesis in 3T3-L1 adipocytes through LKB1/AMPK pathway. PLoS ONE 8, e70135 (2013).
Li, L. et al. Ursolic acid promotes the neuroprotection by activating Nrf2 pathway after cerebral ischemia in mice. Brain Res. 1497, 32–39 (2013).
You, H. J. et al. Ursolic acid enhances nitric oxide and tumor necrosis factor-α production via nuclear factor-κB activation in the resting macrophages. FEBS Lett. 509, 156–160 (2001).
Shishodia, S., Majumdar, S., Banerjee, S. & Aggarwal, B. B. Ursolic acid inhibits nuclear factor-κB activation induced by carcinogenic agents through suppression of IκBα kinase and p65 phosphorylation: correlation with down-regulation of cyclooxygenase 2, matrix metalloproteinase 9, and cyclin D1. Cancer Res. 63, 4375–4383 (2003).
Lin, J. et al. Ursolic acid promotes colorectal cancer cell apoptosis and inhibits cell proliferation via modulation of multiple signaling pathways. Int. J. Oncol. 43, 1235–1243 (2013).
Pathak, A. K. et al. Ursolic acid inhibits STAT3 activation pathway leading to suppression of proliferation and chemosensitization of human multiple myeloma cells. Mol. Cancer Res. 5, 943–955 (2007).
Cha, H. J. et al. Ursolic acid-induced down-regulation of MMP-9 gene is mediated through the nuclear translocation of glucocorticoid receptor in HT1080 human fibrosarcoma cells. Oncogene 16, 771–778 (1998).
Chang, M. R., Lyda, B., Kamenecka, T. M. & Griffin, P. R. Pharmacological repression of RORγ is therapeutic in the collagen induced arthritis experimental model. Arthritis Rheum. http://dx.doi.org/10.1002/art.38272 (2013).
Huang, W. et al. Identification of potent and selective RORγ antagonists. In: Probe Reports from the NIH Molecular Libraries Program [online], (US National Center for Biotechnology Information, 2010).
Zhang, W. et al. Increasing human Th17 differentiation through activation of orphan nuclear receptor retinoid acid-related orphan receptor γ (RORγ) by a class of aryl amide compounds. Mol. Pharmacol. 82, 583–590 (2012).
Acknowledgements
The work of T.P.B. is supported by US National Institutes of Health (NIH) grants MH093429 and MH092769.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
DATABASES
Glossary
- DNA response element
-
A short sequence of DNA that is specifically recognized and bound by a particular transcription factor. DNA response elements are often found in the promoter regions of genes, and confer the responsiveness of a gene to regulation by a particular transcription factor.
- E box
-
A particular DNA response element that is recognized by transcription factors belonging to the basic helix–loop–helix domain-containing family, such as circadian locomotor output cycles protein kaput (CLOCK) and brain and muscle ARNT-like 1 (BMAL1).
- Period (τ)
-
The time that elapses for one complete oscillation or cycle of a particular activity (for example, locomotor activity). Typically, the period for a circadian rhythm is almost 24 hours. In the absence of any extrinsic stimuli that act to 'entrain' the circadian rhythm (such as light), the period may differ; for example, mice typically have a period of slightly less than 24 hours in the absence of entrainment.
- ROR response element
-
A particular DNA response element that is recognized by retinoic acid receptor-related orphan receptors (RORs) and REV-ERBs.
- T helper 17 cells
-
(TH17 cells). A subset of TH cells that produce interleukin-17 (IL-17) and provide microbial immunity at mucosa and epithelial barriers. They have been implicated in the development of autoimmune disease.
- T cell
-
A type of lymphocyte that has a crucial role in cellular immunity. T cells can be distinguished from other lymphocytes based on the expression of the T cell receptor on their plasma membrane.
- Apo structure
-
A receptor structure that is free from a bound ligand.
- Inverse agonists
-
Ligands that suppress the basal activity of a receptor.
- Wheel running activity
-
A measure of locomotor activity as defined by rodents running on a wheel within a cage.
- Phase shift
-
A discrete alteration in an oscillation in locomotor activity or other measurable physiological activity along the time axis within a circadian rhythm.
- Area under the curve
-
(AUC). The area under the curve that is generated by plotting the concentration of a drug in plasma against time.
- Melatonin
-
A hormone that is produced by the pineal gland in a circadian manner and is associated with entrainment of the circadian rhythm.
- Fibroblast growth factor 21
-
(FGF21). A hormone that has several metabolic activities. FGF21 protects animals from diet-induced obesity and lowers blood glucose and lipid levels when administered to diabetic rodents.
- TReg cells
-
A subset of T cells that produce interleukin-10 (IL-10) and transforming growth factor-β (TGFβ) and have an important role in immune tolerance.
- TH1 cells
-
A subset of T helper (TH) cells that produces interferon-γ (IFNγ) and has an important role in cellular immunity.
- TH2 cells
-
A subset of T helper (TH) cells that produces interleukin-4 (IL-4), IL-5 and IL-13, and has an important role in humoral immunity.
Rights and permissions
About this article
Cite this article
Kojetin, D., Burris, T. REV-ERB and ROR nuclear receptors as drug targets. Nat Rev Drug Discov 13, 197–216 (2014). https://doi.org/10.1038/nrd4100
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrd4100
This article is cited by
-
RORα inhibits gastric cancer proliferation through attenuating G6PD and PFKFB3 induced glycolytic activity
Cancer Cell International (2024)
-
Delineating the role of nuclear receptors in colorectal cancer, a focused review
Discover Oncology (2024)
-
Brain nuclear receptors and cardiovascular function
Cell & Bioscience (2023)
-
Targeted screening and identification of chlorhexidine as a pro-myogenic circadian clock activator
Stem Cell Research & Therapy (2023)
-
Cardiac-targeted delivery of nuclear receptor RORα via ultrasound targeted microbubble destruction optimizes the benefits of regular dose of melatonin on sepsis-induced cardiomyopathy
Biomaterials Research (2023)