Key Points
-
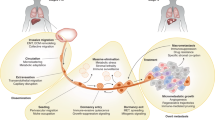
Metastasis is the major cause of morbidity for patients with breast cancer, as few curative therapies are available.
-
To develop more effective treatments, a better understanding of metastasis and the genes that regulate the process is necessary.
-
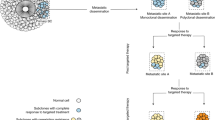
Owing to early tumour cell dissemination before primary tumour diagnosis, target genes need to be identified that have a functional role in metastatic progression after tumour cell entry into the circulation.
-
Current therapies that target the primary tumour may not necessarily target disseminated tumour cells or subsequent metastases. The use of circulating tumour cells to predict and monitor patient response to therapies may be important for improving individualized therapeutics.
-
There are two general ways of identifying metastasis-associated genes as potential therapeutic targets. Human gene expression profiling or tissue arrays of primary tumours, disseminated tumour cells and metastases can be used to find genes whose expression correlates with clinical parameters such as disease-free survival. Assessment of a functional role in the process can be achieved using appropriate animal models of metastatic disease. Alternatively, metastasis-regulating genes can be identified using mouse models and subsequently verified as being relevant in human breast cancer by analysing transcript or protein levels in tissue samples.
-
Approximately 20% of the patients who are diagnosed with breast cancer will subsequently develop metastatic disease. Challenges exist in identifying the patients for whom adjuvant chemotherapy is required. Testing the efficacy of current and emerging therapeutics against disseminated tumour cells in the adjuvant setting is of crucial importance for the future.
Abstract
Nearly all deaths caused by solid cancers occur as a result of metastasis — the formation of secondary tumours in distant organs such as the lungs, liver, brain and bone. A major obstruction to the development of drugs with anti-metastatic efficacy is our fragmented understanding of how tumours 'evolve' and metastasize, at both the biological and genetic levels. Furthermore, although there is significant overlap in the metastatic process among different types of cancer, there are also marked differences in the propensity to metastasize, the extent of metastasis, the sites to which the tumour metastasizes, the kinetics of the process and the mechanisms involved. Here, we consider the case of breast cancer, which has some marked distinguishing features compared with other types of cancer. Considerable progress has been made in the development of preclinical models and in the identification of relevant signalling pathways and genetic regulators of metastatic breast cancer, and we discuss how these might facilitate the development of novel targeted anti-metastatic drugs.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
American Cancer Society. Cancer Facts & Figures 2011. Atlanta: American Cancer Society (2011).
Bernards, R. & Weinberg, R. A. A progression puzzle. Nature 418, 823 (2002).
Nguyen, D. X. & Massague, J. Genetic determinants of cancer metastasis. Nature Rev. Genet. 8, 341–352 (2007).
Kang, Y. et al. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell 3, 537–549 (2003).
Smith, S. C. & Theodorescu, D. Learning therapeutic lessons from metastasis suppressor proteins. Nature Rev. Cancer 9, 253–264 (2009).
Erler, J. T. & Weaver, V. M. Three-dimensional context regulation of metastasis. Clin. Exp. Metastasis 26, 35–49 (2009).
Fidler, I. J., Kim, S. J. & Langley, R. R. The role of the organ microenvironment in the biology and therapy of cancer metastasis. J. Cell Biochem. 101, 927–936 (2007).
Bervar, A. et al. Invasiveness of transformed human breast epithelial cell lines is related to cathepsin B and inhibited by cysteine proteinase inhibitors. Biol. Chem. 384, 447–455 (2003).
Chang, C. & Werb, Z. The many faces of metalloproteases: cell growth, invasion, angiogenesis and metastasis. Trends Cell Biol. 11, S37–S43 (2001).
Liotta, L. A. & Kohn, E. C. The microenvironment of the tumour–host interface. Nature 411, 375–379 (2001).
Caporale, A. et al. Has desmoplastic response extent protective action against tumor aggressiveness in gastric carcinoma? J. Exp. Clin. Cancer Res. 20, 21–24 (2001).
Cardone, A., Tolino, A., Zarcone, R., Borruto Caracciolo, G. & Tartaglia, E. Prognostic value of desmoplastic reaction and lymphocytic infiltration in the management of breast cancer. Panminerva Med. 39, 174–177 (1997).
Iacobuzio-Donahue, C. A., Argani, P., Hempen, P. M., Jones, J. & Kern, S. E. The desmoplastic response to infiltrating breast carcinoma: gene expression at the site of primary invasion and implications for comparisons between tumor types. Cancer Res. 62, 5351–5357 (2002).
Erler, J. T. et al. Hypoxia-induced lysyl oxidase is a critical mediator of bone marrow cell recruitment to form the premetastatic niche. Cancer Cell 15, 35–44 (2009).
Werb, Z. ECM and cell surface proteolysis: regulating cellular ecology. Cell 91, 439–442 (1997).
McCarty, O. J., Mousa, S. A., Bray, P. F. & Konstantopoulos, K. Immobilized platelets support human colon carcinoma cell tethering, rolling, and firm adhesion under dynamic flow conditions. Blood 96, 1789–1797 (2000).
Felding-Habermann, B. et al. Integrin activation controls metastasis in human breast cancer. Proc. Natl Acad. Sci. USA 98, 1853–1858 (2001).
Muller, A. et al. Involvement of chemokine receptors in breast cancer metastasis. Nature 410, 50–56 (2001).
Heyn, C. et al. In vivo magnetic resonance imaging of single cells in mouse brain with optical validation. Magn. Reson. Med. 55, 23–29 (2006).
MacDonald, I. C., Groom, A. C. & Chambers, A. F. Cancer spread and micrometastasis development: quantitative approaches for in vivo models. Bioessays 24, 885–893 (2002).
Meng, S. et al. Circulating tumor cells in patients with breast cancer dormancy. Clin. Cancer Res. 10, 8152–8162 (2004).
Barkan, D. et al. Inhibition of metastatic outgrowth from single dormant tumor cells by targeting the cytoskeleton. Cancer Res. 68, 6241–6250 (2008).
Almog, N. et al. Transcriptional switch of dormant tumors to fast-growing angiogenic phenotype. Cancer Res. 69, 836–844 (2009).
Koebel, C. M. et al. Adaptive immunity maintains occult cancer in an equilibrium state. Nature 450, 903–907 (2007).
Eyles, J. et al. Tumor cells disseminate early, but immunosurveillance limits metastatic outgrowth, in a mouse model of melanoma. J. Clin. Invest. 120, 2030–2039 (2010).
Folkman, J. Role of angiogenesis in tumor growth and metastasis. Semin. Oncol. 29, 15–18 (2002).
Carmeliet, P. & Jain, R. K. Molecular mechanisms and clinical applications of angiogenesis. Nature 473, 298–307 (2011).
Folkman, J. Angiogenesis: an organizing principle for drug discovery? Nature Rev. Drug Discov. 6, 273–286 (2007).
Kerbel, R. & Folkman, J. Clinical translation of angiogenesis inhibitors. Nature Rev. Cancer 2, 727–739 (2002).
Bear, H. D. et al. Bevacizumab added to neoadjuvant chemotherapy for breast cancer. N. Engl. J. Med. 366, 310–320 (2012).
Schneider, B. P. & Sledge, G. W. Jr. Drug insight: VEGF as a therapeutic target for breast cancer. Nature Clin. Pract. Oncol. 4, 181–189 (2007).
Gao, D. et al. Endothelial progenitor cells control the angiogenic switch in mouse lung metastasis. Science 319, 195–198 (2008).
Nolan, D. J. et al. Bone marrow-derived endothelial progenitor cells are a major determinant of nascent tumor neovascularization. Genes Dev. 21, 1546–1558 (2007).
Ruzinova, M. B. et al. Effect of angiogenesis inhibition by Id loss and the contribution of bone-marrow-derived endothelial cells in spontaneous murine tumors. Cancer Cell 4, 277–289 (2003).
Hattori, K. et al. Vascular endothelial growth factor and angiopoietin-1 stimulate postnatal hematopoiesis by recruitment of vasculogenic and hematopoietic stem cells. J. Exp. Med. 193, 1005–1014 (2001).
Banerji, S. et al. LYVE-1, a new homologue of the CD44 glycoprotein, is a lymph-specific receptor for hyaluronan. J. Cell Biol. 144, 789–801 (1999).
Ordonez, N. G. Podoplanin: a novel diagnostic immunohistochemical marker. Adv. Anat. Pathol. 13, 83–88 (2006).
Saharinen, P., Tammela, T., Karkkainen, M. J. & Alitalo, K. Lymphatic vasculature: development, molecular regulation and role in tumor metastasis and inflammation. Trends Immunol. 25, 387–395 (2004).
Nathanson, S. D. Insights into the mechanisms of lymph node metastasis. Cancer 98, 413–423 (2003).
Sundar, S. S. & Ganesan, T. S. Role of lymphangiogenesis in cancer. J. Clin. Oncol. 25, 4298–4307 (2007).
Achen, M. G., Mann, G. B. & Stacker, S. A. Targeting lymphangiogenesis to prevent tumour metastasis. Br. J. Cancer 94, 1355–1360 (2006).
Chang, H. Y. et al. Robustness, scalability, and integration of a wound-response gene expression signature in predicting breast cancer survival. Proc. Natl Acad. Sci. USA 102, 3738–3743 (2005).
Orimo, A. et al. Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell 121, 335–348 (2005).
Olumi, A. F. et al. Carcinoma-associated fibroblasts direct tumor progression of initiated human prostatic epithelium. Cancer Res. 59, 5002–5011 (1999).
Bhowmick, N. A., Neilson, E. G. & Moses, H. L. Stromal fibroblasts in cancer initiation and progression. Nature 432, 332–337 (2004).
Bhowmick, N. A. et al. TGF-beta signaling in fibroblasts modulates the oncogenic potential of adjacent epithelia. Science 303, 848–851 (2004).
Sugimoto, H., Mundel, T. M., Kieran, M. W. & Kalluri, R. Identification of fibroblast heterogeneity in the tumor microenvironment. Cancer Biol. Ther. 5, 1640–1646 (2006).
Grum-Schwensen, B. et al. Suppression of tumor development and metastasis formation in mice lacking the S100A4(mts1) gene. Cancer Res. 65, 3772–3780 (2005).
Hu, M. et al. Distinct epigenetic changes in the stromal cells of breast cancers. Nature Genet. 37, 899–905 (2005).
Karnoub, A. E. et al. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature 449, 557–563 (2007).
Direkze, N. C. et al. Bone marrow contribution to tumor-associated myofibroblasts and fibroblasts. Cancer Res. 64, 8492–8495 (2004).
Bissell, M. J. & Radisky, D. Putting tumours in context. Nature Rev. Cancer 1, 46–54 (2001).
Adriance, M. C., Inman, J. L., Petersen, O. W. & Bissell, M. J. Myoepithelial cells: good fences make good neighbors. Breast Cancer Res. 7, 190–197 (2005).
Coussens, L. M. & Werb, Z. Inflammation and cancer. Nature 420, 860–867 (2002).
Erez, N. & Coussens, L. M. Leukocytes as paracrine regulators of metastasis and determinants of organ-specific colonization. Int. J. Cancer 128, 2536–2544 (2011).
Diaz-Montero, C. M. et al. Increased circulating myeloid-derived suppressor cells correlate with clinical cancer stage, metastatic tumor burden, and doxorubicin-cyclophosphamide chemotherapy. Cancer Immunol. Immunother. 58, 49–59 (2009).
Solito, S. et al. A human promyelocytic-like population is responsible for the immune suppression mediated by myeloid-derived suppressor cells. Blood 118, 2254–2265 (2011).
Sinha, P. et al. Proinflammatory S100 proteins regulate the accumulation of myeloid-derived suppressor cells. J. Immunol. 181, 4666–4675 (2008).
Huang, B. et al. Gr-1+CD115+ immature myeloid suppressor cells mediate the development of tumor-induced T regulatory cells and T-cell anergy in tumor-bearing host. Cancer Res. 66, 1123–1131 (2006).
DuPre, S. A. & Hunter, K. W. Jr. Murine mammary carcinoma 4T1 induces a leukemoid reaction with splenomegaly: association with tumor-derived growth factors. Exp. Mol. Pathol. 82, 12–24 (2007).
Yang, L. et al. Abrogation of TGFβ signaling in mammary carcinomas recruits Gr-1+CD11b+ myeloid cells that promote metastasis. Cancer Cell 13, 23–35 (2008).
Mauti, L. A. et al. Myeloid-derived suppressor cells are implicated in regulating permissiveness for tumor metastasis during mouse gestation. J. Clin. Invest. 121, 2794–2807 (2011).
Bingle, L., Brown, N. J. & Lewis, C. E. The role of tumour-associated macrophages in tumour progression: implications for new anticancer therapies. J. Pathol. 196, 254–265 (2002).
Murdoch, C., Giannoudis, A. & Lewis, C. E. Mechanisms regulating the recruitment of macrophages into hypoxic areas of tumors and other ischemic tissues. Blood 104, 2224–2234 (2004).
Allavena, P., Sica, A., Solinas, G., Porta, C. & Mantovani, A. The inflammatory micro-environment in tumor progression: the role of tumor-associated macrophages. Crit. Rev. Oncol. Hematol. 66, 1–9 (2008).
Locati, M. et al. Analysis of the gene expression profile activated by the CC chemokine ligand 5/RANTES and by lipopolysaccharide in human monocytes. J. Immunol. 168, 3557–3562 (2002).
Lin, E. Y., Nguyen, A. V., Russell, R. G. & Pollard, J. W. Colony-stimulating factor 1 promotes progression of mammary tumors to malignancy. J. Exp. Med. 193, 727–740 (2001).
Burns, C. J. & Wilks, A. F. c-FMS inhibitors: a patent review. Expert Opin. Ther. Pat. 21, 147–165 (2011).
Martin, P. & Leibovich, S. J. Inflammatory cells during wound repair: the good, the bad and the ugly. Trends Cell Biol. 15, 599–607 (2005).
Hiratsuka, S., Watanabe, A., Aburatani, H. & Maru, Y. Tumour-mediated upregulation of chemoattractants and recruitment of myeloid cells predetermines lung metastasis. Nature Cell Biol. 8, 1369–1375 (2006).
Hiratsuka, S. et al. The S100A8-serum amyloid A3-TLR4 paracrine cascade establishes a pre-metastatic phase. Nature Cell Biol. 10, 1349–1355 (2008).
Kaplan, R. N. et al. VEGFR1-positive haematopoietic bone marrow progenitors initiate the pre-metastatic niche. Nature 438, 820–827 (2005).
Olkhanud, P. B. et al. Breast cancer lung metastasis requires expression of chemokine receptor CCR4 and regulatory T cells. Cancer Res. 69, 5996–6004 (2009).
Lyden, D., Welch, D. R. & Psaila, B. (eds) Cancer Metastasis: Biologic Basis and Therapeutics (Cambridge University Press, 2011).
Early Breast Cancer Trialists' Collaborative Group. Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet 365, 1687–1717 (2005).
Iglehart, J. D. & Silver, D. P. Synthetic lethality — a new direction in cancer-drug development. N. Engl. J. Med. 361, 189–191 (2009).
Gelmon, K. A. et al. Olaparib in patients with recurrent high-grade serous or poorly differentiated ovarian carcinoma or triple-negative breast cancer: a Phase 2, multicentre, open-label, non-randomised study. Lancet Oncol. 12, 852–861 (2011).
Ma, W. W. & Adjei, A. A. Novel agents on the horizon for cancer therapy. CA Cancer J. Clin. 59, 111–137 (2009).
de Bono, J. S. & Ashworth, A. Translating cancer research into targeted therapeutics. Nature 467, 543–549 (2010).
Higgins, M. J. & Baselga, J. Breast cancer in 2010: novel targets and therapies for a personalized approach. Nature Rev. Clin. Oncol. 8, 65–66 (2011).
Lin, S. X. et al. Molecular therapy of breast cancer: progress and future directions. Nature Rev. Endocrinol. 6, 485–493 (2010).
Luu, T., Chung, C. & Somlo, G. Combining emerging agents in advanced breast cancer. Oncologist 16, 760–771 (2011).
Welch, D. R. Microarrays bring new insights into understanding of breast cancer metastasis to bone. Breast Cancer Res. 6, 61–64 (2004).
Schardt, J. A. et al. Genomic analysis of single cytokeratin-positive cells from bone marrow reveals early mutational events in breast cancer. Cancer Cell 8, 227–239 (2005).
Poplawski, A. B. et al. Frequent genetic differences between matched primary and metastatic breast cancer provide an approach to identification of biomarkers for disease progression. Eur. J. Hum. Genet. 18, 560–568 (2010).
Colombo, P. E., Milanezi, F., Weigelt, B. & Reis-Filho, J. S. Microarrays in the 2010s: the contribution of microarray-based gene expression profiling to breast cancer classification, prognostication and prediction. Breast Cancer Res. 13, 212 (2011).
Desmedt, C., Ruiz-Garcia, E. & Andre, F. Gene expression predictors in breast cancer: current status, limitations and perspectives. Eur. J. Cancer 44, 2714–2720 (2008).
Zujewski, J. A. & Kamin, L. Trial assessing individualized options for treatment for breast cancer: the TAILORx trial. Future Oncol. 4, 603–610 (2008).
Cardoso, F. et al. Clinical application of the 70-gene profile: the MINDACT trial. J. Clin. Oncol. 26, 729–735 (2008).
Knauer, M. et al. The predictive value of the 70-gene signature for adjuvant chemotherapy in early breast cancer. Breast Cancer Res. Treat. 120, 655–661 (2010).
Albain, K. S. et al. Prognostic and predictive value of the 21-gene recurrence score assay in postmenopausal women with node-positive, oestrogen-receptor-positive breast cancer on chemotherapy: a retrospective analysis of a randomised trial. Lancet Oncol. 11, 55–65 (2010).
Harbeck, N. et al. Enhanced benefit from adjuvant chemotherapy in breast cancer patients classified high-risk according to urokinase-type plasminogen activator (uPA) and plasminogen activator inhibitor type 1 (n = 3424). Cancer Res. 62, 4617–4622 (2002).
Gennari, A. et al. HER2 status and efficacy of adjuvant anthracyclines in early breast cancer: a pooled analysis of randomized trials. J. Natl Cancer Inst. 100, 14–20 (2008).
Pritchard, K. I. et al. HER2 and responsiveness of breast cancer to adjuvant chemotherapy. N. Engl. J. Med. 354, 2103–2111 (2006).
Giuliano, M. et al. Circulating tumor cells as prognostic and predictive markers in metastatic breast cancer patients receiving first-line systemic treatment. Breast Cancer Res. 13, R67 (2011).
Hartkopf, A., Wagner, P., Wallwiener, D., Fehm, T. & Rothmund, R. Changing levels of circulating tumor cells in monitoring chemotherapy response in patients with metastatic breast cancer. Anticancer Res. 31, 979–984 (2011).
Hayes, D. F. et al. Circulating tumor cells at each follow-up time point during therapy of metastatic breast cancer patients predict progression-free and overall survival. Clin. Cancer Res. 12, 4218–4224 (2006).
De Giorgi, U. et al. Circulating tumor cells and [18F]fluorodeoxyglucose positron emission tomography/computed tomography for outcome prediction in metastatic breast cancer. J. Clin. Oncol. 27, 3303–3311 (2009).
Dawood, S. et al. Circulating tumor cells in metastatic breast cancer: from prognostic stratification to modification of the staging system? Cancer 113, 2422–2430 (2008).
Cristofanilli, M. et al. Circulating tumor cells: a novel prognostic factor for newly diagnosed metastatic breast cancer. J. Clin. Oncol. 23, 1420–1430 (2005).
Cristofanilli, M. et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N. Engl. J. Med. 351, 781–791 (2004).
Pachmann, K. et al. Monitoring the response of circulating epithelial tumor cells to adjuvant chemotherapy in breast cancer allows detection of patients at risk of early relapse. J. Clin. Oncol. 26, 1208–1215 (2008).
Pachmann, K. et al. Quantification of the response of circulating epithelial cells to neoadjuvant treatment for breast cancer: a new tool for therapy monitoring. Breast Cancer Res. 7, R975–R979 (2005).
Pierga, J. Y. et al. Circulating tumor cell detection predicts early metastatic relapse after neoadjuvant chemotherapy in large operable and locally advanced breast cancer in a Phase II randomized trial. Clin. Cancer Res. 14, 7004–7010 (2008).
Slade, M. J. et al. Comparison of bone marrow, disseminated tumour cells and blood-circulating tumour cells in breast cancer patients after primary treatment. Br. J. Cancer 100, 160–166 (2009).
Fehm, T. et al. HER2 status of circulating tumor cells in patients with metastatic breast cancer: a prospective, multicenter trial. Breast Cancer Res. Treat. 124, 403–412 (2010).
Wulfing, P. et al. HER2-positive circulating tumor cells indicate poor clinical outcome in stage I to III breast cancer patients. Clin. Cancer Res. 12, 1715–1720 (2006).
Tewes, M. et al. Molecular profiling and predictive value of circulating tumor cells in patients with metastatic breast cancer: an option for monitoring response to breast cancer related therapies. Breast Cancer Res. Treat. 115, 581–590 (2009).
Meng, S. et al. HER-2 gene amplification can be acquired as breast cancer progresses. Proc. Natl Acad. Sci. USA 101, 9393–9398 (2004).
Cabioglu, N. et al. Chemokine receptors in advanced breast cancer: differential expression in metastatic disease sites with diagnostic and therapeutic implications. Ann. Oncol. 20, 1013–1019 (2009).
Zidan, J. et al. Comparison of HER-2 overexpression in primary breast cancer and metastatic sites and its effect on biological targeting therapy of metastatic disease. Br. J. Cancer 93, 552–556 (2005).
Braun, S. et al. Lack of effect of adjuvant chemotherapy on the elimination of single dormant tumor cells in bone marrow of high-risk breast cancer patients. J. Clin. Oncol. 18, 80–86 (2000).
Pantel, K. et al. Differential expression of proliferation-associated molecules in individual micrometastatic carcinoma cells. J. Natl Cancer Inst. 85, 1419–1424 (1993).
Muller, V., Alix-Panabieres, C. & Pantel, K. Insights into minimal residual disease in cancer patients: implications for anti-cancer therapies. Eur. J. Cancer 46, 1189–1197 (2010).
Fehm, T., Muller, V., Alix-Panabieres, C. & Pantel, K. Micrometastatic spread in breast cancer: detection, molecular characterization and clinical relevance. Breast Cancer Res. 10, S1 (2008).
Aktas, B. et al. Comparison of estrogen and progesterone receptor status of circulating tumor cells and the primary tumor in metastatic breast cancer patients. Gynecol. Oncol. 122, 356–360 (2011).
Folkman, J., Merler, E., Abernathy, C. & Williams, G. Isolation of a tumor factor responsible for angiogenesis. J. Exp. Med. 133, 275–288 (1971).
Bergers, G. & Hanahan, D. Modes of resistance to anti-angiogenic therapy. Nature Rev. Cancer 8, 592–603 (2008).
Ebos, J. M. et al. Accelerated metastasis after short-term treatment with a potent inhibitor of tumor angiogenesis. Cancer Cell 15, 232–239 (2009).
Paez-Ribes, M. et al. Antiangiogenic therapy elicits malignant progression of tumors to increased local invasion and distant metastasis. Cancer Cell 15, 220–231 (2009).
Schneider, B. P. & Sledge, G. W. Jr. Anti-VEGF therapy as adjuvant therapy: clouds on the horizon? Breast Cancer Res. 11, 303 (2009).
Loges, S., Mazzone, M., Hohensinner, P. & Carmeliet, P. Silencing or fueling metastasis with VEGF inhibitors: antiangiogenesis revisited. Cancer Cell 15, 167–170 (2009).
Kerbel, R. S. Tumor angiogenesis. N. Engl. J. Med. 358, 2039–2049 (2008).
Shojaei, F. et al. Tumor refractoriness to anti-VEGF treatment is mediated by CD11b+Gr1+ myeloid cells. Nature Biotech. 25, 911–920 (2007).
Ahn, G. O. & Brown, J. M. Matrix metalloproteinase-9 is required for tumor vasculogenesis but not for angiogenesis: role of bone marrow-derived myelomonocytic cells. Cancer Cell 13, 193–205 (2008).
Quesada, A. R., Medina, M. A. & Alba, E. Playing only one instrument may be not enough: limitations and future of the antiangiogenic treatment of cancer. Bioessays 29, 1159–1168 (2007).
Nguyen, D. X., Bos, P. D. & Massague, J. Metastasis: from dissemination to organ-specific colonization. Nature Rev. Cancer 9, 274–284 (2009).
Henderson, M. A. et al. Parathyroid hormone-related protein localization in breast cancers predict improved prognosis. Cancer Res. 66, 2250–2256 (2006).
Fleming, N. I. et al. Parathyroid hormone-related protein protects against mammary tumor emergence and is associated with monocyte infiltration in ductal carcinoma in situ. Cancer Res. 69, 7473–7479 (2009).
Riethdorf, S., Wikman, H. & Pantel, K. Review: biological relevance of disseminated tumor cells in cancer patients. Int. J. Cancer 123, 1991–2006 (2008).
Pantel, K., Brakenhoff, R. H. & Brandt, B. Detection, clinical relevance and specific biological properties of disseminating tumour cells. Nature Rev. Cancer 8, 329–340 (2008).
Ignatiadis, M., Georgoulias, V. & Mavroudis, D. Micrometastatic disease in breast cancer: clinical implications. Eur. J. Cancer 44, 2726–2736 (2008).
Watson, M. A. et al. Isolation and molecular profiling of bone marrow micrometastases identifies TWIST1 as a marker of early tumor relapse in breast cancer patients. Clin. Cancer Res. 13, 5001–5009 (2007).
Lorger, M., Lee, H., Forsyth, J. S. & Felding-Habermann, B. Comparison of in vitro and in vivo approaches to studying brain colonization by breast cancer cells. J. Neurooncol. 104, 689–696 (2011).
Arguello, F., Baggs, R. B. & Frantz, C. N. A murine model of experimental metastasis to bone and bone marrow. Cancer Res. 48, 6876–6881 (1988).
Luzzi, K. J. et al. Multistep nature of metastatic inefficiency: dormancy of solitary cells after successful extravasation and limited survival of early micrometastases. Am. J. Pathol. 153, 865–873 (1998).
Shaeffer, J., El-Mahdi, A. M. & Constable, W. C. An experimental model for the treatment of pulmonary metastases. Eur. J. Cancer 11, 523–525 (1975).
Sharpless, N. E. & Depinho, R. A. The mighty mouse: genetically engineered mouse models in cancer drug development. Nature Rev. Drug Discov. 5, 741–754 (2006).
Tsukamoto, A. S., Grosschedl, R., Guzman, R. C., Parslow, T. & Varmus, H. E. Expression of the int-1 gene in transgenic mice is associated with mammary gland hyperplasia and adenocarcinomas in male and female mice. Cell 55, 619–625 (1988).
Jeffers, M. et al. The mutationally activated Met receptor mediates motility and metastasis. Proc. Natl Acad. Sci. USA 95, 14417–14422 (1998).
Derksen, P. W. et al. Somatic inactivation of E-cadherin and p53 in mice leads to metastatic lobular mammary carcinoma through induction of anoikis resistance and angiogenesis. Cancer Cell 10, 437–449 (2006).
Shepard, C. R., Kassis, J., Whaley, D. L., Kim, H. G. & Wells, A. PLCγ contributes to metastasis of in situ-occurring mammary and prostate tumors. Oncogene 26, 3020–3026 (2007).
Yeh, E. S. et al. Hunk is required for HER2/neu-induced mammary tumorigenesis. J. Clin. Invest. 121, 866–879 (2011).
Canaani, D. Methodological approaches in application of synthetic lethality screening towards anticancer therapy. Br. J. Cancer 100, 1213–1218 (2009).
Varticovski, L. et al. Accelerated preclinical testing using transplanted tumors from genetically engineered mouse breast cancer models. Clin. Cancer Res. 13, 2168–2177 (2007).
Coleman, R. E. & Rubens, R. D. The clinical course of bone metastases from breast cancer. Br. J. Cancer 55, 61–66 (1987).
Dexter, D. L. et al. Heterogeneity of tumor cells from a single mouse mammary tumor. Cancer Res. 38, 3174–3181 (1978).
Hiraga, T. et al. Effects of oral UFT combined with or without zoledronic acid on bone metastasis in the 4T1/luc mouse breast cancer. Int. J. Cancer 106, 973–979 (2003).
Lelekakis, M. et al. A novel orthotopic model of breast cancer metastasis to bone. Clin. Exp. Metastasis 17, 163–170 (1999).
Eckhardt, B. L. et al. Genomic analysis of a spontaneous model of breast cancer metastasis to bone reveals a role for the extracellular matrix. Mol. Cancer Res. 3, 1–13 (2005).
Rose, A. A. et al. Osteoactivin promotes breast cancer metastasis to bone. Mol. Cancer Res. 5, 1001–1014 (2007).
Ewens, A., Mihich, E. & Ehrke, M. J. Distant metastasis from subcutaneously grown E0771 medullary breast adenocarcinoma. Anticancer Res. 25, 3905–3915 (2005).
Brown, G. B., Bendich, A., Roll, P. M. & Kanematsu, S. Utilization of guanine by the C57 black mouse bearing adenocarcinoma Eo771. Proc. Soc. Exp. Biol. Med. 72, 501–502 (1949).
Duss, S. et al. An oestrogen-dependent model of breast cancer created by transformation of normal human mammary epithelial cells. Breast Cancer Res. 9, R38 (2007).
Lev, D. C., Kiriakova, G. & Price, J. E. Selection of more aggressive variants of the gI101A human breast cancer cell line: a model for analyzing the metastatic phenotype of breast cancer. Clin. Exp. Metastasis 20, 515–523 (2003).
Bandyopadhyay, A. et al. A soluble transforming growth factor β type III receptor suppresses tumorigenicity and metastasis of human breast cancer MDA-MB-231 cells. Cancer Res. 59, 5041–5046 (1999).
Wang, H. et al. IL-12 gene-modified bone marrow cell therapy suppresses the development of experimental metastatic prostate cancer. Cancer Gene Ther. 14, 819–827 (2007).
Kuperwasser, C. et al. A mouse model of human breast cancer metastasis to human bone. Cancer Res. 65, 6130–6138 (2005).
Minn., A. J. et al. Genes that mediate breast cancer metastasis to lung. Nature 436, 518–524 (2005).
Lam, P. et al. A human bone NOD/SCID mouse model to distinguish metastatic potential in primary breast cancers. Cancer Biol. Ther. 8, 1010–1017 (2009).
Beckhove, P. et al. Efficient engraftment of human primary breast cancer transplants in nonconditioned NOD/Scid mice. Int. J. Cancer 105, 444–453 (2003).
DeRose, Y. S. et al. Tumor grafts derived from women with breast cancer authentically reflect tumor pathology, growth, metastasis and disease outcomes. Nature Med. 17, 1514–1520 (2011).
O'Neill, K., Lyons, S. K., Gallagher, W. M., Curran, K. M. & Byrne, A. T. Bioluminescent imaging: a critical tool in pre-clinical oncology research. J. Pathol. 220, 317–327 (2010).
Hoffman, R. M. The multiple uses of fluorescent proteins to visualize cancer in vivo. Nature Rev. Cancer 5, 796–806 (2005).
Filonov, G. S. et al. Bright and stable near-infrared fluorescent protein for in vivo imaging. Nature Biotech. 29, 757–761 (2011).
Solomon, B., McArthur, G., Cullinane, C., Zalcberg, J. & Hicks, R. Applications of positron emission tomography in the development of molecular targeted cancer therapeutics. BioDrugs 17, 339–354 (2003).
Beer, A. J. & Schwaiger, M. Imaging of integrin αvβ3 expression. Cancer Metastasis Rev. 27, 631–644 (2008).
Palmieri, D. et al. Brain metastases of breast cancer. Breast Dis. 26, 139–147 (2006).
Sameni, M. et al. Imaging and quantifying the dynamics of tumor-associated proteolysis. Clin. Exp. Metastasis 26, 299–309 (2009).
Withana, N. P. et al. Cathepsin B inhibition limits bone metastasis in breast cancer. Cancer Res. 72, 1199–1209 (2012).
Schena, M., Shalon, D., Davis, R. W. & Brown, P. O. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270, 467–470 (1995).
Sotiriou, C. et al. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J. Natl Cancer Inst. 98, 262–272 (2006).
van 't Veer, L. J. et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature 415, 530–536 (2002).
Buffa, F. M. et al. MicroRNA-associated progression pathways and potential therapeutic targets identified by integrated mRNA and microRNA expression profiling in breast cancer. Cancer Res. 71, 5635–5645 (2011).
Sorlie, T. et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl Acad. Sci. USA 98, 10869–10874 (2001).
Gruvberger, S. et al. Estrogen receptor status in breast cancer is associated with remarkably distinct gene expression patterns. Cancer Res. 61, 5979–5984 (2001).
Ahr, A. et al. Identification of high risk breast-cancer patients by gene expression profiling. Lancet 359, 131–132 (2002).
Hedenfalk, I. et al. Gene-expression profiles in hereditary breast cancer. N. Engl. J. Med. 344, 539–548 (2001).
Cimino, A. et al. Epithelial cell adhesion molecule (EpCAM) is overexpressed in breast cancer metastases. Breast Cancer Res. Treat. 123, 701–708 (2010).
Steeg, P. S., Ouatas, T., Halverson, D., Palmieri, D. & Salerno, M. Metastasis suppressor genes: basic biology and potential clinical use. Clin. Breast Cancer 4, 51–62 (2003).
Hurst, D. R., Edmonds, M. D. & Welch, D. R. Metastamir: the field of metastasis-regulatory microRNA is spreading. Cancer Res. 69, 7495–7498 (2009).
Yang, J. et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell 117, 927–939 (2004).
Andrews, J. et al. Multi-platform whole-genome microarray analyses refine the epigenetic signature of breast cancer metastasis with gene expression and copy number. PLoS One 5, e8665 (2010).
Hu, G. et al. MTDH activation by 8q22 genomic gain promotes chemoresistance and metastasis of poor-prognosis breast cancer. Cancer Cell 15, 9–20 (2009).
Fang, F. et al. Breast cancer methylomes establish an epigenomic foundation for metastasis. Sci. Transl. Med. 3, 75ra25 (2011).
Yan, L. X. et al. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA 14, 2348–2360 (2008).
Brown, D. M. & Ruoslahti, E. Metadherin, a cell surface protein in breast tumors that mediates lung metastasis. Cancer Cell 5, 365–374 (2004).
Bodenstine, T. M. & Welch, D. R. Metastasis suppressors and the tumor microenvironment. Cancer Microenviron. 1, 1–11 (2008).
Kauffman, E. C., Robinson, V. L., Stadler, W. M., Sokoloff, M. H. & Rinker-Schaeffer, C. W. Metastasis suppression: the evolving role of metastasis suppressor genes for regulating cancer cell growth at the secondary site. J. Urol. 169, 1122–1133 (2003).
Gupta, P. B. et al. The melanocyte differentiation program predisposes to metastasis after neoplastic transformation. Nature Genet. 37, 1047–1054 (2005).
Jechlinger, M. et al. Autocrine PDGFR signaling promotes mammary cancer metastasis. J. Clin. Invest. 116, 1561–1570 (2006).
Waerner, T. et al. ILEI: a cytokine essential for EMT, tumor formation, and late events in metastasis in epithelial cells. Cancer Cell 10, 227–239 (2006).
Cowin, P. & Welch, D. R. Breast cancer progression: controversies and consensus in the molecular mechanisms of metastasis and EMT. J. Mammary Gland Biol. Neoplasia 12, 99–102 (2007).
Kalluri, R. & Weinberg, R. A. The basics of epithelial–mesenchymal transition. J. Clin. Invest. 119, 1420–1428 (2009).
Huber, M. A., Kraut, N. & Beug, H. Molecular requirements for epithelial–mesenchymal transition during tumor progression. Curr. Opin. Cell Biol. 17, 548–558 (2005).
Thiery, J. P. Epithelial–mesenchymal transitions in tumour progression. Nature Rev. Cancer 2, 442–454 (2002).
Thiery, J. P. Epithelial–mesenchymal transitions in development and pathologies. Curr. Opin. Cell Biol. 15, 740–746 (2003).
Hugo, H. et al. Epithelial–mesenchymal and mesenchymal–epithelial transitions in carcinoma progression. J. Cell Physiol. 213, 374–383 (2007).
Lee, J. M., Dedhar, S., Kalluri, R. & Thompson, E. W. The epithelial–mesenchymal transition: new insights in signaling, development, and disease. J. Cell Biol. 172, 973–981 (2006).
Bos, P. D. et al. Genes that mediate breast cancer metastasis to the brain. Nature 459, 1005–1009 (2009).
Wang, Y. et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet 365, 671–679 (2005).
Ramaswamy, S., Ross, K. N., Lander, E. S. & Golub, T. R. A molecular signature of metastasis in primary solid tumors. Nature Genet. 33, 49–54 (2003).
Wirapati, P. et al. Meta-analysis of gene expression profiles in breast cancer: toward a unified understanding of breast cancer subtyping and prognosis signatures. Breast Cancer Res. 10, R65 (2008).
Thakur, D. et al. Microproteomic analysis of 10,000 laser captured microdissected breast tumor cells using short-range sodium dodecyl sulfate-polyacrylamide gel electrophoresis and porous layer open tubular liquid chromatography tandem mass spectrometry. J. Chromatogr. A 1218, 8168–8174 (2011).
Pasqualini, R. & Ruoslahti, E. Organ targeting in vivo using phage display peptide libraries. Nature 380, 364–366 (1996).
Kolonin, M. G. et al. Ligand-directed surface profiling of human cancer cells with combinatorial peptide libraries. Cancer Res. 66, 34–40 (2006).
Gupta, R. A. et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464, 1071–1076 (2010).
Gumireddy, K. et al. KLF17 is a negative regulator of epithelial–mesenchymal transition and metastasis in breast cancer. Nature Cell Biol. 11, 1297–1304 (2009).
Gumireddy, K. et al. In vivo selection for metastasis promoting genes in the mouse. Proc. Natl Acad. Sci. USA 104, 6696–6701 (2007).
Pravtcheva, D. D. & Wise, T. L. Metastasizing mammary carcinomas in H19 enhancers-Igf2 transgenic mice. J. Exp. Zool. 281, 43–57 (1998).
Teuliere, J. et al. Targeted activation of β-catenin signaling in basal mammary epithelial cells affects mammary development and leads to hyperplasia. Development 132, 267–277 (2005).
Guy, C. T. et al. Expression of the neu protooncogene in the mammary epithelium of transgenic mice induces metastatic disease. Proc. Natl Acad. Sci. USA 89, 10578–10582 (1992).
Guy, C. T., Cardiff, R. D. & Muller, W. J. Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol. Cell Biol. 12, 954–961 (1992).
Kitsberg, D. I. & Leder, P. Keratinocyte growth factor induces mammary and prostatic hyperplasia and mammary adenocarcinoma in transgenic mice. Oncogene 13, 2507–2515 (1996).
Liu, C. H. et al. Overexpression of cyclooxygenase-2 is sufficient to induce tumorigenesis in transgenic mice. J. Biol. Chem. 276, 18563–18569 (2001).
D'Cruz, C. M. et al. c-MYC induces mammary tumorigenesis by means of a preferred pathway involving spontaneous Kras2 mutations. Nature Med. 7, 235–239 (2001).
Kwan, H. et al. Transgenes expressing the Wnt-1 and int-2 proto-oncogenes cooperate during mammary carcinogenesis in doubly transgenic mice. Mol. Cell Biol. 12, 147–154 (1992).
Zinser, G. M. et al. Mammary-specific Ron receptor overexpression induces highly metastatic mammary tumors associated with β-catenin activation. Cancer Res. 66, 11967–11974 (2006).
Kwak, E. L. et al. Mammary tumorigenesis following transgenic expression of a dominant negative CHK2 mutant. Cancer Res. 66, 1923–1928 (2006).
Nielsen, L. L., Discafani, C. M., Gurnani, M. & Tyler, R. D. Histopathology of salivary and mammary gland tumors in transgenic mice expressing a human Ha-ras oncogene. Cancer Res. 51, 3762–3767 (1991).
Gallego, M. I., Bierie, B. & Hennighausen, L. Targeted expression of HGF/SF in mouse mammary epithelium leads to metastatic adenosquamous carcinomas through the activation of multiple signal transduction pathways. Oncogene 22, 8498–8508 (2003).
Gallahan, D. et al. Expression of a truncated Int3 gene in developing secretory mammary epithelium specifically retards lobular differentiation resulting in tumorigenesis. Cancer Res. 56, 1775–1785 (1996).
Sloan, E. K. et al. Tumor-specific expression of αvβ3 integrin promotes spontaneous metastasis of breast cancer to bone. Breast Cancer Res. 8, R20 (2006).
Eckhardt, B. L., Parker, B. S., Restall, C. M., Van Laar, R. K. & Anderson, R. L. Identification and characterization of novel genetic regulators of breast cancer metastasis. Clin. Exp. Metastasis 24, 299–300 (2007).
Shevde, L. A. et al. Suppression of human melanoma metastasis by the metastasis suppressor gene, BRMS1. Exp. Cell Res. 273, 229–239 (2002).
Seraj, M. J., Samant, R. S., Verderame, M. F. & Welch, D. R. Functional evidence for a novel human breast carcinoma metastasis suppressor, BRMS1, encoded at chromosome 11q13. Cancer Res. 60, 2764–2769 (2000).
Samant, R. S. et al. Analysis of mechanisms underlying BRMS1 suppression of metastasis. Clin. Exp. Metastasis 18, 683–693 (2000).
Lopez, J. I. et al. CD44 attenuates metastatic invasion during breast cancer progression. Cancer Res. 65, 6755–6763 (2005).
Yang, X. et al. Overexpression of KAI1 suppresses in vitro invasiveness and in vivo metastasis in breast cancer cells. Cancer Res. 61, 5284–5288 (2001).
Dong, J. T. et al. KAI1, a metastasis suppressor gene for prostate cancer on human chromosome 11p11.2. Science 268, 884–886 (1995).
Takaoka, A. et al. Reduced invasive and metastatic potentials of KAI1-transfected melanoma cells. Jpn J. Cancer Res. 89, 397–404 (1998).
Jiang, Y. et al. KiSS1 suppresses metastasis in human ovarian cancer via inhibition of protein kinase C alpha. Clin. Exp. Metastasis 22, 369–376 (2005).
Lee, J. H. et al. KiSS-1, a novel human malignant melanoma metastasis-suppressor gene. J. Natl Cancer Inst. 88, 1731–1737 (1996).
Lee, J. H. & Welch, D. R. Suppression of metastasis in human breast carcinoma MDA-MB-435 cells after transfection with the metastasis suppressor gene, KiSS-1. Cancer Res. 57, 2384–2387 (1997).
Leone, A. et al. Reduced tumor incidence, metastatic potential, and cytokine responsiveness of nm23-transfected melanoma cells. Cell 65, 25–35 (1991).
Leone, A., Flatow, U., VanHoutte, K. & Steeg, P. S. Transfection of human nm23-H1 into the human MDA-MB-435 breast carcinoma cell line: effects on tumor metastatic potential, colonization and enzymatic activity. Oncogene 8, 2325–2333 (1993).
Tagashira, H., Hamazaki, K., Tanaka, N., Gao, C. & Namba, M. Reduced metastatic potential and c-myc overexpression of colon adenocarcinoma cells (Colon 26 line) transfected with nm23–R2/rat nucleoside diphosphate kinase alpha isoform. Int. J. Mol. Med. 2, 65–68 (1998).
Miyazaki, H. et al. Overexpression of nm23-H2/NDP kinase B in a human oral squamous cell carcinoma cell line results in reduced metastasis, differentiated phenotype in the metastatic site, and growth factor-independent proliferative activity in culture. Clin. Cancer Res. 5, 4301–4307 (1999).
Li, H. Z. et al. Effects of Raf kinase inhibitor protein expression on metastasis and progression of human breast cancer. Mol. Cancer Res. 7, 832–840 (2009).
Fu, Z. et al. Effects of Raf kinase inhibitor protein expression on suppression of prostate cancer metastasis. J. Natl Cancer Inst. 95, 878–889 (2003).
Parker, B. S. et al. Primary tumour expression of the cysteine cathepsin inhibitor stefin A inhibits distant metastasis in breast cancer. J. Pathol. 214, 337–346 (2008).
Sevenich, L. et al. Transgenic expression of human cathepsin B promotes progression and metastasis of polyoma-middle-T-induced breast cancer in mice. Oncogene 30, 54–64 (2011).
Shimo, T. et al. Pathogenic role of connective tissue growth factor (CTGF/CCN2) in osteolytic metastasis of breast cancer. J. Bone Miner. Res. 21, 1045–1059 (2006).
Lin, B. R. et al. Connective tissue growth factor inhibits metastasis and acts as an independent prognostic marker in colorectal cancer. Gastroenterology 128, 9–23 (2005).
Chang, C. C. et al. Connective tissue growth factor and its role in lung adenocarcinoma invasion and metastasis. J. Natl Cancer Inst. 96, 364–375 (2004).
Aikawa, T., Gunn, J., Spong, S. M., Klaus, S. J. & Korc, M. Connective tissue growth factor-specific antibody attenuates tumor growth, metastasis, and angiogenesis in an orthotopic mouse model of pancreatic cancer. Mol. Cancer Ther. 5, 1108–1116 (2006).
Dornhofer, N. et al. Connective tissue growth factor-specific monoclonal antibody therapy inhibits pancreatic tumor growth and metastasis. Cancer Res. 66, 5816–5827 (2006).
Deckers, M. et al. The tumor suppressor Smad4 is required for transforming growth factor β-induced epithelial to mesenchymal transition and bone metastasis of breast cancer cells. Cancer Res. 66, 2202–2209 (2006).
Acknowledgements
Fellowship support from the National Health and Medical Research Council of Australia (B.S.P.), from the National Breast Cancer Foundation (R.L.A.) and from Susan G. Komen for the Cure (B.L.E.) is gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
During the past 2 years, Prudence Francis has received travel support from Sanofi and Amgen.
Supplementary information
Supplementary information Table 1
The identification of subtypes of breast cancer with different levels of risk of metastasis. (PDF 128 kb)
Supplementary information Table 2
Current therapies for breast cancer (PDF 141 kb)
Related links
Rights and permissions
About this article
Cite this article
Eckhardt, B., Francis, P., Parker, B. et al. Strategies for the discovery and development of therapies for metastatic breast cancer. Nat Rev Drug Discov 11, 479–497 (2012). https://doi.org/10.1038/nrd2372
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrd2372
This article is cited by
-
Construction of Metastasis-Specific Regulation Network in Ovarian Cancer Based on Prognostic Stemness-Related Signatures
Reproductive Sciences (2023)
-
Huaier polysaccharides suppress triple-negative breast cancer metastasis and epithelial-mesenchymal transition by inducing autophagic degradation of Snail
Cell & Bioscience (2021)
-
Ruyiping formula inhibits metastasis via the microRNA-134-SLUG axis in breast cancer
BMC Complementary Medicine and Therapies (2021)
-
Bone marrow niches in the regulation of bone metastasis
British Journal of Cancer (2021)
-
USP19 modulates cancer cell migration and invasion and acts as a novel prognostic marker in patients with early breast cancer
Oncogenesis (2021)