Key Points
-
The need to analyse large numbers of clinically well-defined tissue specimens constitutes a major bottleneck in target validation.
-
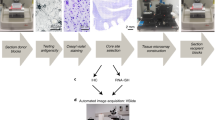
The tissue microarray (TMA) technology allows the simultaneous analysis of thousands of tissue samples in a single experiment. TMAs are, therefore, cost efficient and offer an unprecedented degree of standardization because all tissue samples are subjected to exactly the same experimental conditions and batches of reagents.
-
TMAs can be made from virtually all kinds of diseased and non-diseased tissues, including formalin-fixed and fresh frozen tissues, xenograft tissues and cell lines.
-
Numerous in situ analysis methods can be used on TMA sections, including immunohistochemistry, fluorescence in situ hybridization (FISH) and RNA in situ hybridization.
-
Despite the small size of arrayed samples (diameter 0.6 mm), TMA studies provide highly representative information of the donor tissues.
-
TMAs accelerate the process of drug discovery at several key steps, including validation of drug targets and determination of their molecular epidemiology, estimation of potential treatment-related side effects and the development of diagnostic assays.
-
The TMA format is optimally suited for the storage and economical follow-up analysis of limited tissue resources, for example, tissues collected from clinical studies.
-
Automation of the reading of stained TMA sections, and standards for TMA database development and management, will be key issues in the future.
Abstract
Advances in molecular methods have massively facilitated the discovery of potential molecular targets for gene-specific therapy. Accelerated lead discovery has at the same time generated a massive demand for thorough validation of such putative targets. Very often human tissue analysis is needed for this purpose. However, the need to analyse large numbers of well-characterized human tissues constitutes a major bottleneck in drug discovery and development. Traditional tissue analysis in a slide-by-slide manner is slow, expensive and difficult to standardize. In addition, precious specimens, such as tissue samples from clinical studies, are usually exhausted after a few analyses. The tissue microarray technology overcomes these shortcomings as it allows the simultaneous analysis of up to 1,000 minute tissue samples in a single experiment. This article will review how high-throughput tissue microarray analyses can dramatically facilitate translational research at several different levels.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Goldstine, J., Seligson, D. B., Beizai, P., Miyata, H. & Vinters, H. V. Tissue microarrays in the study of non-neoplastic disease of the nervous system. J. Neuropathol. Exp. Neurol. 61, 653–662 (2002).
Bubendorf, L. et al. Survey of gene amplifications during prostate cancer progression by high-throughput fluorescence in situ hybridization on tissue microarrays. Cancer Res. 59, 803–806 (1999).
Kononen, J. et al. Tissue microarrays for high-throughput molecular profiling of hundreds of specimens. Nature Med. 4, 844–847 (1998). Initial publication of the TMA technology.
Fejzo, M. S. & Slamon, D. J. Frozen tumor tissue microarray technology for analysis of tumor RNA, DNA, and proteins. Am. J. Pathol. 159, 1645–1650 (2001). Describes TMAs from frozen tissue samples.
Simon, R. & Sauter, G. Tissue microarrays for miniaturized high-throughput molecular profiling of tumors. Exp. Hematol. 30, 1365–1372 (2002).
Gancberg, D. et al. Reliability of the tissue microarray based FISH for evaluation of the HER-2 oncogene in breast carcinoma. J. Clin. Pathol. 55, 315–317 (2002).
Camp, R. L., Charette, L. A. & Rimm, D. L. Validation of tissue microarray technology in breast carcinoma. Lab. Invest. 80, 1943–1949 (2000). An early TMA-validation study.
Rimm, D. L. et al. Tissue microarray: a new technology for amplification of tissue resources. Cancer J. 7, 24–31 (2001).
Gancberg, D. et al. Evaluation of HER-2/NEU protein expression in breast cancer by immunohistochemistry: an interlaboratory study assessing the reproducibility of HER-2/NEU testing. Breast Cancer Res. Treat. 74, 113–120 (2002).
Ginestier, C. et al. Distinct and complementary information provided by use of tissue and DNA microarrays in the study of breast tumor markers. Am. J. Pathol. 161, 1223–1233 (2002).
Torhorst, J. et al. Tissue microarrays for rapid linking of molecular changes to clinical endpoints. Am. J. Pathol. 159, 2249–2256 (2001).
Hendriks, Y. et al. Conventional and tissue microarray immunohistochemical expression analysis of mismatch repair in hereditary colorectal tumors. Am. J. Pathol. 162, 469–477 (2003).
Fernebro, E., Dictor, M., Bendahl, P. O., Ferno, M. & Nilbert, M. Evaluation of the tissue microarray technique for immunohistochemical analysis in rectal cancer. Arch. Pathol. Lab. Med. 126, 702–705 (2002).
Mucci, N. R., Akdas, G., Manely, S. & Rubin, M. A. Neuroendocrine expression in metastatic prostate cancer: evaluation of high throughput tissue microarrays to detect heterogeneous protein expression. Hum. Pathol. 31, 406–414 (2000).
Rubin, M. A., Dunn, R., Strawderman, M. & Pienta, K. J. Tissue microarray sampling strategy for prostate cancer biomarker analysis. Am. J. Surg. Pathol. 26, 312–319 (2002). Illustrates that four punches per tissue sample provide optimal representativity.
Merseburger, A. S. et al. Limitations of tissue microarrays in the evaluation of focal alterations of bcl-2 and p53 in whole mount derived prostate tissues. Oncol. Rep. 10, 223–228 (2003).
Gulmann, C., Butler, D., Kay, E., Grace, A. & Leader, M. Biopsy of a biopsy: validation of immunoprofiling in gastric cancer biopsy tissue microarrays. Histopathology 42, 70–76 (2003).
Moch, H. et al. High-throughput tissue microarray analysis to evaluate genes uncovered by cDNA microarray screening in renal cell carcinoma. Am. J. Pathol. 154, 981–986 (1999).
Nocito, A. et al. Microarrays of bladder cancer tissue are highly representative of proliferation index and histological grade. J. Pathol. 194, 349–357 (2001).
Sallinen, S. L. et al. Identification of differentially expressed genes in human gliomas by DNA microarray and tissue chip techniques. Cancer Res. 60, 6617–6622 (2000).
Engellau, J. et al. Tissue microarray technique in soft tissue sarcoma: immunohistochemical Ki-67 expression in malignant fibrous histiocytoma. Appl. Immunohistochem. Mol. Morphol. 9, 358–363 (2001).
Hoos, A. et al. Validation of tissue microarrays for immunohistochemical profiling of cancer specimens using the example of human fibroblastic tumors. Am. J. Pathol. 158, 1245–1251 (2001).
Rassidakis, G. Z. et al. Apoptotic rate in peripheral T-cell lymphomas. A study using a tissue microarray with validation on full tissue sections. Am. J. Clin. Pathol. 118, 328–334 (2002).
Garcia, J. F. et al. Hodgkin and Reed-Sternberg cells harbor alterations in the major tumor suppressor pathways and cell-cycle checkpoints: analyses using tissue microarrays. Blood 101, 681–689 (2003). Reports TMA of Hodgkin's lymphoma samples.
Hedvat, C. V. et al. Application of tissue microarray technology to the study of non-Hodgkin's and Hodgkin's lymphoma. Hum. Pathol. 33, 968–974 (2002). Describes TMA of Hodgkin's lymphoma samples.
Tzankov, A. et al. High-throughput tissue microarray analysis of G1-cyclin alterations in classical Hodgkin's lymphoma indicates overexpression of cyclin E1. J. Pathol. 199, 201–207 (2003). An analysis of TMA of Hodgkin's lymphoma samples.
Torhorst, J. et al. Tissue microarrays for rapid linking of molecular changes to clinical endpoints. Am. J. Pathol. 159, 2249–2256 (2001). Reports TMA evaluation of known prognostic factors (ER,PR) in breast cancer.
Barlund, M. et al. Detecting activation of ribosomal protein S6 kinase by complementary DNA and tissue microarray analysis. J. Natl Cancer Inst. 92, 1252–1259 (2000). This study combines cDNA and TMA technologies.
Hoos, A. et al. High Ki-67 proliferative index predicts disease specific survival in patients with high-risk soft tissue sarcomas. Cancer 92, 869–874 (2001).
Hoos, A. et al. Clinical significance of molecular expression profiles of Hurthle cell tumors of the thyroid gland analyzed via tissue microarrays. Am. J. Pathol. 160, 175–183 (2002).
Bubendorf, L. et al. Hormone therapy failure in human prostate cancer: analysis by complementary DNA and tissue microarrays. J. Natl Cancer Inst. 91, 1758–1764 (1999). Describes a combination of cDNA and TMA technologies.
Dhanasekaran, S. M. et al. Delineation of prognostic biomarkers in prostate cancer. Nature 412, 822–826 (2001).
Porkka, K., Saramaki, O., Tanner, M. & Visakorpi, T. Amplification and overexpression of Elongin C gene discovered in prostate cancer by cDNA microarrays. Lab. Invest. 82, 629–637 (2002).
Mousses, S. et al. Clinical validation of candidate genes associated with prostate cancer progression in the CWR22 model system using tissue microarrays. Cancer Res. 62, 1256–1260 (2002).
Xin, W., Rhodes, D. R., Ingold, C., Chinnaiyan, A. M. & Rubin, M. A. Dysregulation of the annexin family protein family is associated with prostate cancer progression. Am. J. Pathol. 162, 255–261 (2003).
Otsuka, M. et al. Differential expression of the L-plastin gene in human colorectal cancer progression and metastasis. Biochem. Biophys. Res. Commun. 289, 876–881 (2001).
Nielsen, T. O. et al. Molecular characterisation of soft tissue tumours: a gene expression study. Lancet 359, 1301–1307 (2002).
Allander, S. V. et al. Expression profiling of synovial sarcoma by cDNA microarrays: association of ERBB2, IGFBP2, and ELF3 with epithelial differentiation. Am. J. Pathol. 161, 1587–1595 (2002).
Wasenius, V. M. et al. Hepatocyte growth factor receptor, matrix metalloproteinase-11, tissue inhibitor of metalloproteinase-1, and fibronectin are up-regulated in papillary thyroid carcinoma: a cDNA and tissue microarray study. Clin. Cancer Res. 9, 68–75 (2003).
Oka, T. et al. Reduction of hematopoietic cell-specific tyrosine phosphatase SHP-1 gene expression in natural killer cell lymphoma and various types of lymphomas/ leukemias: combination analysis with cDNA expression array and tissue microarray. Am. J. Pathol. 159, 1495–1505 (2001).
Heiskanen, M. et al. CGH, cDNA and tissue microarray analyses implicate FGFR2 amplification in a small subset of breast tumors. Anal. Cell. Pathol. 22, 229–234 (2001).
Hyman, E. et al. Impact of DNA amplification on gene expression patterns in breast cancer. Cancer Res. 62, 6240–6245 (2002). CGH on cDNA microarrays.
Richter, J. et al. High-throughput tissue microarray analysis of cyclin E gene amplification and overexpression in urinary bladder cancer. Am. J. Pathol. 157, 787–794 (2000). Describes an example of a prognosis TMA.
Simon, R. et al. High-throughput tissue microarray analysis of 3p25 (RAF1) and 8p12 (FGFR1) copy number alterations in urinary bladder cancer. Cancer Res. 61, 4514–4519 (2001).
Simon, R. et al. Amplification pattern of 12q13–q15 genes (MDM2, CDK4, GLI) in urinary bladder cancer. Oncogene 21, 2476–2483 (2002). Amplicon mapping using different TMAs.
Kocher, T. et al. Prognostic relevance of MAGE-A4 tumor antigen expression in transitional cell carcinoma of the urinary bladder: a tissue microarray study. Int. J. Cancer 100, 702–705 (2002).
Park, S. Y., Kim, H. S., Hong, E. K. & Kim, W. H. Expression of cytokeratins 7 and 20 in primary carcinomas of the stomach and colorectum and their value in the differential diagnosis of metastatic carcinomas to the ovary. Hum. Pathol. 33, 1078–1085 (2002).
Saramaki, O. et al. Amplification of EIF3S3 gene is associated with advanced stage in prostate cancer. Am. J. Pathol. 159, 2089–2094 (2001).
Skacel, M. et al. Aneusomy of chromosomes 7, 8, and 17 and amplification of HER-2/neu and epidermal growth factor receptor in Gleason score 7 prostate carcinoma: a differential fluorescent in situ hybridization study of Gleason pattern 3 and 4 using tissue microarray. Hum. Pathol. 32, 1392–1397 (2001).
Rubin, M. A. et al. E-cadherin expression in prostate cancer: a broad survey using high-density tissue microarray technology. Hum. Pathol. 32, 690–697 (2001).
Fuller, C. E., Wang, H., Zhang, W., Fuller, G. N. & Perry, A. High-throughput molecular profiling of high-grade astrocytomas: the utility of fluorescence in situ hybridization on tissue microarrays (TMA-FISH). J. Neuropathol. Exp. Neurol. 61, 1078–1084 (2002).
Ristimaki, A. et al. Prognostic significance of elevated cyclooxygenase-2 expression in breast cancer. Cancer Res. 62, 632–635 (2002).
Miettinen, H. E. et al. High topoisomerase II α expression associates with high proliferation rate and and poor prognosis in oligodendrogliomas. Neuropathol. Appl. Neurobiol. 26, 504–512 (2000).
Wang, Y. et al. Prognostic significance of c-myc and AIB1 amplification in hepatocellular carcinoma. A broad survey using high-throughput tissue microarray. Cancer 95, 2346–2352 (2002).
Simon, R. et al. Patterns of her-2/neu amplification and overexpression in primary and metastatic breast cancer. J. Natl Cancer Inst. 93, 1141–1146 (2001). This paper provides an example of a metastasis TMA.
Maloney, D. G. et al. IDEC-C2B8 (Rituximab) anti-CD20 monoclonal antibody therapy in patients with relapsed low-grade non-Hodgkin's lymphoma. Blood 90, 2188–2195 (1997).
Rao, J., Seligson, D. & Hemstreet, G. P. Protein expression analysis using quantitative fluorescence image analysis on tissue microarray slides. Biotechniques 32, 924–926, 928–930, 932 (2002).
Camp, R. L., Chung, G. G. & Rimm, D. L. Automated subcellular localization and quantification of protein expression in tissue microarrays. Nature Med. 8, 1323–1327 (2002). Describes an automated TMA analysis using fluorescent dyes.
Haedicke, W., Popper, H. H., Buck, C. R. & Zatloukal, K. Automated evaluation and normalization of immunohistochemistry on tissue microarrays with a DNA microarray scanner. Biotechniques 35, 164–168 (2003). Report of an automated TMA analysis.
Liu, C. L. et al. Software tools for high-throughput analysis and archiving of immunohistochemistry staining data obtained with tissue microarrays. Am. J. Pathol. 161, 1557–1565 (2002).
Berman, J. J., Edgerton, M. E. & Friedman, B. A. The tissue microarray data exchange specification: A community-based, open source tool for sharing tissue microarray data. BMC Med. Inform. Decis. Mak. 3, 5 (2003).
Schraml, P. et al. Tissue microarrays for gene amplification surveys in many different tumor types. Clin. Cancer Res. 5, 1966–1975 (1999). Provides an example of a multitumour TMA.
Sauer, T., Wiedswang, G., Boudjema, G., Christensen, H. & Karesen, R. Assessment of HER-2/neu overexpression and/or gene amplification in breast carcinomas: should in situ hybridization be the method of choice? APMIS 111, 444–450 (2003).
Zhang, D., Salto-Tellez, M., Do, E., Putti, T. C. & Koay, E. S. Evaluation of HER-2/neu oncogene status in breast tumors on tissue microarrays. Hum. Pathol. 34, 362–368 (2003).
Latta, E. K., Tjan, S., Parkes, R. K. & O'Malley, F. P. The role of HER2/neu overexpression/amplification in the progression of ductal carcinoma in situ to invasive carcinoma of the breast. Mod. Pathol. 15, 1318–1325 (2002).
Bozzetti, C. et al. HER-2/neu amplification detected by fluorescence in situ hybridization in fine needle aspirates from primary breast cancer. Ann. Oncol. 13, 1398–1403 (2002).
Tsuda, H. et al. Detection of HER-2/neu (c-erb B-2) DNA amplification in primary breast carcinoma. Interobserver reproducibility and correlation with immunohistochemical HER-2 overexpression. Cancer 92, 2965–2974 (2001).
Riou, G. et al. c-erbB-2 (HER-2/neu) gene amplification is a better indicator of poor prognosis than protein over-expression in operable breast-cancer patients. Int. J. Cancer 95, 266–270 (2001).
Chen, Y., Dong, J. & Li, C. Amplification and overexpression of c-erbB2 in human breast cancer. Zhonghua Zhong Liu Za Zhi 17, 16–19 (1995).
Hubbard, A. L., Doris, C. P., Thompson, A. M., Chetty, U. & Anderson, T. J. Critical determination of the frequency of c-erbB-2 amplification in breast cancer. Br. J. Cancer 70, 434–439 (1994).
Pechoux, C., Chardonnet, Y. & Noel, P. Immunohistochemical studies on c-erbB-2 oncoprotein expression in paraffin embedded tissues in invasive and non-invasive human breast lesions. Anticancer Res. 14, 1343–1360 (1994).
Bermont, L., Algros, M. P., Baron, M. H. & Adessi, G. L. Relevance of p185 HER-2/neu oncoprotein quantification in human primary breast carcinoma. Breast Cancer Res. Treat. 63, 163–169 (2000).
Knyazev, P. G., Imyanitov, E. N., Chernitca, O. I., Nikiforova, I. F. & Hanson, K. P. Amplification of ERBB-2 (HER-2/NEU) oncogene in different neoplasms of patients from USSR. Oncology 49, 162–165 (1992).
Fajac, A. et al. c-erbB2 gene amplification and protein expression in ovarian epithelial tumors: evaluation of their respective prognostic significance by multivariate analysis. Int. J. Cancer 64, 146–151 (1995).
Fan, Q. B. et al. Amplification of the C-erbB-2(HER-2/neu) proto-oncogene in ovarian carcinomas. Chin. Med. J. (Engl) 107, 589–593 (1994).
Hou, Y., Wang, M. & Liu, W. Study of the amplification and expression of c-erbB2 oncogene in epithelial ovarian tumors. Zhonghua Zhong Liu Za Zhi 18, 426–428 (1996).
Anreder, M. B., Freeman, S. M., Merogi, A., Halabi, S. & Marrogi, A. J. p53, c-erbB2, and PCNA status in benign, proliferative and malignant ovarian surface epithelial neoplasms: a study of 75 cases. Arch. Pathol. Lab. Med. 123, 310–316 (1999).
Berchuck, A. et al. Overexpression of HER-2/neu is associated with poor survival in advanced epithelial ovarian cancer. Cancer Res. 50, 4087–4091 (1990).
Huettner, P. C. et al. Neu oncogene expression in ovarian tumors: a quantitative study. Mod. Pathol. 5, 250–256 (1992).
Yazici, H., Dolapcioglu, K., Buyru, F. & Dalay, N. Utility of c-erbB-2 expression in tissue and sera of ovarian cancer patients. Cancer Invest. 18, 110–114 (2000).
Haldane, J. S., Hird, V., Hughes, C. M. & Gullick, W. J. c-erbB-2 oncogene expression in ovarian cancer. J. Pathol. 162, 231–237 (1990).
Wang, D. P. et al. Immunohistochemical localization of c-erbB-2 protein and epidermal growth factor receptor in normal surface epithelium, surface inclusion cysts, and common epithelial tumours of the ovary. Virchows Arch. A Pathol. Anat. Histopathol. 421, 393–400 (1992).
Ross, J. S. et al. HER-2/neu oncogene amplification by fluorescence in situ hybridization in epithelial tumors of the ovary. Am. J. Clin. Pathol. 111, 311–316 (1999).
Wang, Y. P., Chen, S. Q. & Xue, K. X. Amplification of C-erB-2 oncogene in colon carcinomas. Zhonghua Yi Xue Za Zhi 74, 536–538, 582 (1994).
Nathanson, D. R. et al. HER 2/neu expression and gene amplification in colon cancer. Int. J. Cancer 105, 796–802 (2003).
McKay, J. A. et al. c-erbB-2 is not a major factor in the development of colorectal cancer. Br. J. Cancer 86, 568–573 (2002).
Dursun, A., Poyraz, A., Suer, O., Sezer, C. & Akyol, G. Expression of Bcl-2 and c-ErbB-2 in colorectal neoplasia. Pathol. Oncol. Res. 7, 24–27 (2001).
Ross, J. S. & McKenna, B. J. The HER-2/neu oncogene in tumors of the gastrointestinal tract. Cancer Invest. 19, 554–568 (2001).
Esteller, M., Garcia, A., Martinez i Palones, J. M., Cabero, A. & Reventos, J. Detection of c-erbB-2/neu and fibroblast growth factor-3/INT-2 but not epidermal growth factor receptor gene amplification in endometrial cancer by differential polymerase chain reaction. Cancer 75, 2139–2146 (1995).
Czerwenka, K., Lu, Y. & Heuss, F. Amplification and expression of the c-erbB-2 oncogene in normal, hyperplastic, and malignant endometria. Int. J. Gynecol. Pathol. 14, 98–106 (1995).
Halperin, R. et al. Comparative immunohistochemical study of endometrioid and serous papillary carcinoma of endometrium. Eur. J. Gynaecol. Oncol. 22, 122–126 (2001).
Ioffe, O. B., Papadimitriou, J. C. & Drachenberg, C. B. Correlation of proliferation indices, apoptosis, and related oncogene expression (bcl-2 and c-erbB-2) and p53 in proliferative, hyperplastic, and malignant endometrium. Hum. Pathol. 29, 1150–1159 (1998).
Miyazaki, M. Immunohistochemical study of PCNA, p53 gene product and c-erbB-2 gene product in endometrial carcinoma. Nippon Sanka Fujinka Gakkai Zasshi 48, 269–276 (1996).
Rolitsky, C. D., Theil, K. S., McGaughy, V. R., Copeland, L. J. & Niemann, T. H. HER-2/neu amplification and overexpression in endometrial carcinoma. Int. J. Gynecol. Pathol. 18, 138–143 (1999).
Wang, D. et al. Expression of c-erbB-2 protein and epidermal growth receptor in endometrial carcinomas. Correlation with clinicopathologic and sex steroid receptor status. Cancer 72, 2628–2637 (1993).
Pinto-de-Sousa, J. et al. c-erb B-2 expression is associated with tumor location and venous invasion and influences survival of patients with gastric carcinoma. Int. J. Surg. Pathol. 10, 247–256 (2002).
Kameda, T. et al. Expression of ERBB2 in human gastric carcinomas: relationship between p185ERBB2 expression and the gene amplification. Cancer Res. 50, 8002–8009 (1990).
Ranzani, G. N. et al. Heterogeneous protooncogene amplification correlates with tumor progression and presence of metastases in gastric cancer patients. Cancer Res. 50, 7811–7814 (1990).
Tsujino, T. et al. Alterations of oncogenes in metastatic tumours of human gastric carcinomas. Br. J. Cancer 62, 226–230 (1990).
Lee, K. E. et al. Prognostic significance of p53, nm23, PCNA and c-erbB-2 in gastric cancer. Jpn J. Clin. Oncol. 33, 173–179 (2003).
Wang, Y. L., Sheu, B. S., Yang, H. B., Lin, P. W. & Chang, Y. C. Overexpression of c-erb-B2 proteins in tumor and non-tumor parts of gastric adenocarcinoma — emphasis on its relation to H. pylori infection and clinicohistological characteristics. Hepatogastroenterology 49, 1172–1176 (2002).
Takehana, T. et al. Status of c-erbB-2 in gastric adenocarcinoma: a comparative study of immunohistochemistry, fluorescence in situ hybridization and enzyme-linked immuno-sorbent assay. Int. J. Cancer 98, 833–837 (2002).
Suzuki, T. et al. Growth of human gastric carcinomas and expression of epidermal growth factor, transforming growth factor-α, epidermal growth factor receptor and p185c-erbB-2. Oncology 52, 385–391 (1995).
Yan, J. et al. Absence of evidence for HER2 amplification in nasopharyngeal carcinoma. Cancer Genet. Cytogenet. 132, 116–119 (2002).
Khan, A. J. et al. Characterization of the HER-2/neu oncogene by immunohistochemical and fluorescence in situ hybridization analysis in oral and oropharyngeal squamous cell carcinoma. Clin. Cancer Res. 8, 540–548 (2002).
Leonard, J. H., Kearsley, J. H., Chenevix-Trench, G. & Hayward, N. K. Analysis of gene amplification in head-and-neck squamous-cell carcinoma. Int. J. Cancer 48, 511–515 (1991).
Merritt, W. D., Weissler, M. C., Turk, B. F. & Gilmer, T. M. Oncogene amplification in squamous cell carcinoma of the head and neck. Arch. Otolaryngol. Head Neck Surg. 116, 1394–1398 (1990).
Beckhardt, R. N. et al. HER-2/neu oncogene characterization in head and neck squamous cell carcinoma. Arch. Otolaryngol. Head Neck Surg. 121, 1265–1270 (1995).
Yazici, H., Altun, M., Alatli, C., Dogan, O. & Dalay, N. c-erbB-2 gene amplification in nasopharyngeal carcinoma. Cancer Invest. 18, 6–10 (2000).
Cho, K. J., Khang, S. K., Koh, J. S., Chung, J. H. & Lee, S. S. Sebaceous carcinoma of the eyelids: frequent expression of c-erbB-2 oncoprotein. J. Korean Med. Sci. 15, 545–550 (2000).
Shnayder, Y. et al. Adhesion molecules as prognostic factors in nasopharyngeal carcinoma. Laryngoscope 111, 1842–1846 (2001).
Skalova, A., Starek, Kucerova, V., Szepe, P. & Plank, L. Salivary duct carcinoma — a highly aggressive salivary gland tumor with HER-2/neu oncoprotein overexpression. Pathol. Res. Pract. 197, 621–626 (2001).
Shiraishi, M., Noguchi, M., Shimosato, Y. & Sekiya, T. Amplification of protooncogenes in surgical specimens of human lung carcinomas. Cancer Res. 49, 6474–6479 (1989).
Bongiorno, P. F. et al. Alterations of K-ras, p53, and erbB-2/neu in human lung adenocarcinomas. J. Thorac. Cardiovasc. Surg. 107, 590–595 (1994).
Han, H. et al. Prognostic value of immunohistochemical expressions of p53, HER-2/neu, and bcl-2 in stage I non-small-cell lung cancer. Hum. Pathol. 33, 105–110 (2002).
Nakamura, H. et al. Correlation between encoded protein overexpression and copy number of the HER2 gene with survival in non-small cell lung cancer. Int. J. Cancer 103, 61–66 (2003).
Hirsch, F. R. et al. Evaluation of HER-2/neu gene amplification and protein expression in non-small cell lung carcinomas. Br. J. Cancer 86, 1449–1456 (2002).
Hirashima, N., Takahashi, W., Yoshii, S., Yamane, T. & Ooi, A. Protein overexpression and gene amplification of c-erb B-2 in pulmonary carcinomas: a comparative immuno-histochemical and fluorescence in situ hybridization study. Mod. Pathol. 14, 556–562 (2001).
Cox, G. et al. Herceptest: HER2 expression and gene amplification in non-small cell lung cancer. Int. J. Cancer 92, 480–483 (2001).
Reinmuth, N. et al. Ploidy, expression of erbB1, erbB2, P53 and amplification of erbB1, erbB2 and erbB3 in non-small cell lung cancer. Eur. Respir. J. 16, 991–996 (2000).
Brabender, J. et al. Epidermal growth factor receptor and HER2-neu mRNA expression in non-small cell lung cancer is correlated with survival. Clin. Cancer Res. 7, 1850–1855 (2001).
Schneider, P. M. et al. Multiple molecular marker testing (p53, C-Ki-ras, c-erbB-2) improves estimation of prognosis in potentially curative resected non-small cell lung cancer. Br. J. Cancer 83, 473–479 (2000).
Fournier, G. et al. Gene amplifications in advanced-stage human prostate cancer. Urol. Res. 22, 343–347 (1995).
Latil, A. et al. Oncogene amplifications in early-stage human prostate carcinomas. Int. J. Cancer 59, 637–638 (1994).
Ross, J. S. et al. HER-2/neu gene amplification status in prostate cancer by fluorescence in situ hybridization. Hum. Pathol. 28, 827–833 (1997).
Sanchez, K. M. et al. Evaluation of HER-2/neu expression in prostatic adenocarcinoma: a request for a standardized, organ specific methodology. Cancer 95, 1650–1655 (2002).
Lara, P. N. et al. HER-2/neu is overexpressed infrequently in patients with prostate carcinoma. Results from the California Cancer Consortium Screening Trial. Cancer 94, 2584–2589 (2002).
Savinainen, K. J. et al. Expression and gene copy number analysis of ERBB2 oncogene in prostate cancer. Am. J. Pathol. 160, 339–345 (2002).
Liu, H. L., Gandour-Edwards, R., Lara, P. N., Jr., de Vere White, R. & LaSalle, J. M. Detection of low level HER-2/neu gene amplification in prostate cancer by fluorescence in situ hybridization. Cancer J. 7, 395–403 (2001).
Osman, I. et al. HER-2/neu (p185neu) protein expression in the natural or treated history of prostate cancer. Clin. Cancer Res. 7, 2643–2647 (2001).
Gu, K., Mes-Masson, A. M., Gauthier, J. & Saad, F. Overexpression of her-2/neu in human prostate cancer and benign hyperplasia. Cancer Lett. 99, 185–189 (1996).
Shi, Y. et al. Her-2/neu expression in prostate cancer: high level of expression associated with exposure to hormone therapy and androgen independent disease. J. Urol. 166, 1514–1519 (2001).
McCann, A., Dervan, P. A., Johnston, P. A., Gullick, W. J. & Carney, D. N. c-erbB-2 oncoprotein expression in primary human tumors. Cancer 65, 88–92 (1990).
Kruger, S. et al. Overexpression of c-erbB-2 oncoprotein in muscle-invasive bladder carcinoma: relationship with gene amplification, clinicopathological parameters and prognostic outcome. Int. J. Oncol. 21, 981–987 (2002).
Mellon, J. K. et al. C-erbB-2 in bladder cancer: molecular biology, correlation with epidermal growth factor receptors and prognostic value. J. Urol. 155, 321–326 (1996).
Sauter, G. et al. DNA aberrations in urinary bladder cancer detected by flow cytometry and FISH. Urol. Res. 25, S37–S44 (1997).
Gorgoulis, V. G., Barbatis, C., Poulias, I. & Karameris, A. M. Molecular and immunohistochemical evaluation of epidermal growth factor receptor and c-erb-B-2 gene product in transitional cell carcinomas of the urinary bladder: a study in Greek patients. Mod. Pathol. 8, 758–764 (1995).
Orlando, C. et al. Detection of c-erbB-2 amplification in transitional cell bladder carcinoma using competitive PCR technique. J. Urol. 156, 2089–2093 (1996).
Simon, R. et al. HER-2 and TOP2A co-amplification in urinary bladder cancer. Int. J. Cancer 107, 764–772 (2003).
Zhau, H. E. et al. Amplification and expression of the c-erb B-2/neu proto-oncogene in human bladder cancer. Mol. Carcinog. 3, 254–257 (1990).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Guido Sauter has a patent application on the use of tissue microarrays.
Related links
Related links
DATABASES
LocusLink
fibroblast growth factor receptor-1
FURTHER INFORMATION
Institute of Pathology, University Hospital Basel, Tissue Microarray Program
National Human Genome Research Insitute's Tissue Microarray Project
http://www.nhgri.nih.gov/DIR/CGB/TMA/construction.html
Manufacturers of tissue arrayers
Automated TMA analysis systems and software
Software programs for TMA image storage and retrieval
Stanford Tissue Microarray Software
Glossary
- MICROTOME
-
Device for cutting histological sections from tissue blocks.
- IMMUNOHISTOCHEMISTRY
-
(IHC). Method for the in situ detection of proteins in tissues using a specific antibody coupled to a chromogenic enzyme complex. The staining allows a rough estimation of the expression level and the intracellular localization of the target protein.
- FLUORESCENCE IN SITU HYBRIDIZATION (FISH).
-
Method for the detection of particular DNA sequences (for example, a gene) in cell nuclei, for example, in tissue sections. A fluorescence- labelled DNA fragment complementary to the target DNA sequence is used as a probe. FISH allows the determination of the exact copy number of a target gene.
- RNA IN SITU HYBRIDIZATION (RNA-ISH).
-
Method for the detection of mRNA sequences in tissue sections. A (usually isotopic) labelled DNA or RNA fragment complementary to the target mRNA sequence is used as a probe. The signal intensity allows a rough estimation of the mRNA expression level.
- KI67 LABELLING INDEX
-
The Ki67 labelling index is the fraction of cell nuclei positive for staining with an antibody against the Ki67 protein. The Ki67 antigen is exclusively expressed during cell cycle.
- NORTHERN BLOT
-
Method for the detection of mRNA sequences in isolated total cellular RNA that has been separated according to the mRNA length using an isotopic labelled antisense DNA or RNA probe. The signal intensity allows a rough estimation of the mRNA expression level, whereas the signal localizations enables to estimated the target mRNA size.
- PHOSPHORIMAGER
-
Device for visualization of radioactive signals originating from experiments such as isotopic RNA-ISH or Northern blots, comparable with a radiosensitive photographic plate.
Rights and permissions
About this article
Cite this article
Sauter, G., Simon, R. & Hillan, K. Tissue microarrays in drug discovery. Nat Rev Drug Discov 2, 962–972 (2003). https://doi.org/10.1038/nrd1254
Issue Date:
DOI: https://doi.org/10.1038/nrd1254
This article is cited by
-
CUL4B-DDB1-COP1-mediated UTX downregulation promotes colorectal cancer progression
Experimental Hematology & Oncology (2023)
-
High density of CXCL12-positive immune cell infiltration predicts chemosensitivity and recurrence-free survival in ovarian carcinoma
Journal of Cancer Research and Clinical Oncology (2023)
-
The prognostic significance of CXCR4 and SDF-1 in differentiated thyroid cancer depends on CD8+ density
BMC Endocrine Disorders (2022)
-
Prognostic significance of CD8+ T-cells density in stage III colorectal cancer depends on SDF-1 expression
Scientific Reports (2021)
-
High density of CD66b in primary high-grade ovarian cancer independently predicts response to chemotherapy
Journal of Cancer Research and Clinical Oncology (2020)