Abstract
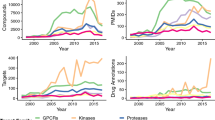
A large proportion of biomedical research and the development of therapeutics is focused on a small fraction of the human genome. In a strategic effort to map the knowledge gaps around proteins encoded by the human genome and to promote the exploration of currently understudied, but potentially druggable, proteins, the US National Institutes of Health launched the Illuminating the Druggable Genome (IDG) initiative in 2014. In this article, we discuss how the systematic collection and processing of a wide array of genomic, proteomic, chemical and disease-related resource data by the IDG Knowledge Management Center have enabled the development of evidence-based criteria for tracking the target development level (TDL) of human proteins, which indicates a substantial knowledge deficit for approximately one out of three proteins in the human proteome. We then present spotlights on the TDL categories as well as key drug target classes, including G protein-coupled receptors, protein kinases and ion channels, which illustrate the nature of the unexplored opportunities for biomedical research and therapeutic development.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Change history
23 March 2018
In the version of this article that was originally published online, an older version of the data set categorizing proteins into target development levels was used to create Figure 1 than the version used to create Table 1, and data from Figure 1 were referred to at several points in the text of the article. Figure 1 and the associated text have been updated to match Table 1 in the online and print versions of the article. The authors apologize for any inconvenience that this error may have caused.
References
Knowles, J. & Gromo, G. Target selection in drug discovery. Nat. Rev. Drug Discov. 2, 63–69 (2003).
Edwards, A. M. et al. Too many roads not taken. Nature 470, 163–165 (2011).
Alberts, B., Kirschner, M. W., Tilghman, S. & Varmus, H. Rescuing US biomedical research from its systemic flaws. Proc. Natl Acad. Sci. USA 111, 5773–5777 (2014).
Kim, S. et al. PubChem Substance and Compound databases. Nucleic Acids Res. 44, D1202–D1213 (2016).
Gaulton, A. et al. The ChEMBL database in 2017. Nucleic Acids Res. 45, D945–D954 (2017).
Tomczak, K., Czerwin´ska, P. & Wiznerowicz, M. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp. Oncol. 19, A68–A77 (2015).
Munafò, M. R. et al. A manifesto for reproducible science. Nat. Hum. Behav. 1, 0021 (2017).
Nickerson, R. S. Confirmation bias: a ubiquitous phenomenon in many guises. Rev. Gen. Psychol. 2, 175–220 (1998).
Santos, R. et al. A comprehensive map of molecular drug targets. Nat. Rev. Drug Discov. 16, 19–34 (2017).
Ursu, O. et al. DrugCentral: online drug compendium. Nucleic Acids Res. 45, D932–D939 (2017).
Amberger, J., Bocchini, C. A., Scott, A. F. & Hamosh, A. McKusick's Online Mendelian Inheritance in Man (OMIM). Nucleic Acids Res. 37, D793–D796 (2009).
Ashburner, M. et al. Gene ontology: tool for the unification of biology. Nat. Genet. 25, 25–29 (2000).
Pletscher-Frankild, S., Pallejà, A., Tsafou, K., Binder, J. X. & Jensen, L. J. Diseases: text mining and data integration of disease-gene associations. Methods 74, 83–89 (2015).
Kiermer, V. Antibodypedia. Nat. Methods 5, 860–861 (2008).
UniProt Consortium. UniProt: a hub for protein information. Nucleic Acids Res. 43, D204–D212 (2015).
Papadatos, G. et al. SureChEMBL: a large-scale, chemically annotated patent document database. Nucleic Acids Res. 44, D1220–1228 (2016).
Rouillard, A. D. et al. The harmonizome: a collection of processed datasets gathered to serve and mine knowledge about genes and proteins. Database 2016, baw100 (2016).
Szklarczyk, D. et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 43, D447–D452 (2015).
Hajduk, P. J., Huth, J. R. & Tse, C. Predicting protein druggability. Drug Discov. Today 10, 1675–1682 (2005).
Hopkins, A. L. & Groom, C. R. The druggable genome. Nat. Rev. Drug Discov. 1, 727–730 (2002).
Surade, S. & Blundell, T. L. Structural biology and drug discovery of difficult targets: the limits of ligandability. Chem. Biol. 19, 42–50 (2012).
Kubinyi, H. Drug research: myths, hype and reality. Nat. Rev. Drug Discov. 2, 665–668 (2003).
Yang, X. et al. Widespread expansion of protein interaction capabilities by alternative splicing. Cell 164, 805–817 (2016).
Mestres, J., Gregori-Puigjané, E., Valverde, S. & Solé, R. V. Data completeness—the Achilles heel of drug-target networks. Nat. Biotechnol. 26, 983–984 (2008).
Schreiber, S. L. et al. Advancing biological understanding and therapeutics discovery with small-molecule probes. Cell 161, 1252–1265 (2015).
Austin, C. P., Brady, L. S., Insel, T. R. & Collins, F. S. NIH molecular libraries initiative. Science 306, 1138–1139 (2004).
Southan, C. et al. The IUPHAR/BPS guide to pharmacology in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. Nucleic Acids Res. 44, D1054–D1068 (2016).
Waring, M. J. et al. An analysis of the attrition of drug candidates from four major pharmaceutical companies. Nat. Rev. Drug Discov. 14, 475–486 (2015).
Hunter, S. et al. InterPro in 2011: new developments in the family and domain prediction database. Nucleic Acids Res. 40, D306–312 (2012).
Kruger, F. A., Gaulton, A., Nowotka, M. & Overington, J. P. PPDMs-a resource for mapping small molecule bioactivities from ChEMBL to Pfam-A protein domains. Bioinformatics 31, 776–778 (2015).
Campillos, M., Kuhn, M., Gavin, A.-C., Jensen, L. J. & Bork, P. Drug target identification using side-effect similarity. Science 321, 263–266 (2008).
Keiser, M. J. et al. Predicting new molecular targets for known drugs. Nature 462, 175–181 (2009).
Huang, X. & Dixit, V. M. Drugging the undruggables: exploring the ubiquitin system for drug development. Cell Res. 26, 484–498 (2016).
Lai, A. C. & Crews, C. M. Induced protein degradation: an emerging drug discovery paradigm. Nat. Rev. Drug Discov. 16, 101–114 (2017).
Sakamoto, K. M. et al. Protacs: chimeric molecules that target proteins to the Skp1–Cullin–F box complex for ubiquitination and degradation. Proc. Natl Acad. Sci. 98, 8554–8559 (2001).
Gadd, M. S. et al. Structural basis of PROTAC cooperative recognition for selective protein degradation. Nat. Chem. Biol. 13, 514–521 (2017).
Mungall, C. J. et al. The Monarch Initiative: an integrative data and analytic platform connecting phenotypes to genotypes across species. Nucleic Acids Res. 45, D712–D722 (2017).
Dickinson, M. E. et al. High-throughput discovery of novel developmental phenotypes. Nature 537, 508–514 (2016).
MacArthur, J. et al. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res. 45, D896–D901 (2017).
GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 45, 580–585 (2013).
GTEx Consortium et al. Genetic effects on gene expression across human tissues. Nature 550, 204–213 (2017).
Uhlén, M. et al. Proteomics. Tissue-based map of the human proteome. Science 347, 1260419 (2015).
Kim, M.-S. et al. A draft map of the human proteome. Nature 509, 575–581 (2014).
Lenat, D. B. & Feigenbaum, E. A. On the thresholds of knowledge. Artif. Intell. 47, 185–250 (1991).
Fishilevich, S. et al. Genic insights from integrated human proteomics in GeneCards. Database 2016, baw030 (2016).
Smirnov, D. A. et al. Genetic variation in radiation-induced cell death. Genome Res. 22, 332–339 (2012).
Garrison, J. L. & Knight, Z. A. Linking smell to metabolism and aging. Science 358, 718–719 (2017).
Kliewer, S. A., Lehmann, J. M. & Willson, T. M. Orphan nuclear receptors: shifting endocrinology into reverse. Science 284, 757–760 (1999).
Willson, T. M., Jones, S. A., Moore, J. T. & Kliewer, S. A. Chemical genomics: functional analysis of orphan nuclear receptors in the regulation of bile acid metabolism. Med. Res. Rev. 21, 513–522 (2001).
Moore, L. B. et al. Orphan nuclear receptors constitutive androstane receptor and pregnane X receptor share xenobiotic and steroid ligands. J. Biol. Chem. 275, 15122–15127 (2000).
Pellicciari, R. et al. 6alpha-ethyl-chenodeoxycholic acid (6-ECDCA), a potent and selective FXR agonist endowed with anticholestatic activity. J. Med. Chem. 45, 3569–3572 (2002).
Hambruch, E., Kinzel, O. & Kremoser, C. On the pharmacology of farnesoid X receptor agonists: give me an 'A', like in 'acid'. Nucl. Recep. Res. 3, 101207 (2016).
Wacker, D., Stevens, R. C. & Roth, B. L. How ligands illuminate GPCR molecular pharmacology. Cell 170, 414–427 (2017).
Roth, B. L., Irwin, J. J. & Shoichet, B. K. Discovery of new GPCR ligands to illuminate new biology. Nat. Chem. Biol. 13, 1143–1151 (2017).
Roth, B. L., Sheffler, D. J. & Kroeze, W. K. Magic shotguns versus magic bullets: selectively non-selective drugs for mood disorders and schizophrenia. Nat. Rev. Drug Discov. 3, 353–359 (2004).
Hernandez, P. A. et al. Mutations in the chemokine receptor gene CXCR4 are associated with WHIM syndrome, a combined immunodeficiency disease. Nat. Genet. 34, 70–74 (2003).
Sternini, C. Receptors and transmission in the brain-gut axis: potential for novel therapies. III. Mu-opioid receptors in the enteric nervous system. Am. J. Physiol. Gastrointest. Liver Physiol. 281, G8–15 (2001).
Sternini, C. Taste receptors in the gastrointestinal tract. IV. Functional implications of bitter taste receptors in gastrointestinal chemosensing. Am. J. Physiol. Gastrointest. Liver Physiol. 292, G457–461 (2007).
Rockman, H. A., Koch, W. J. & Lefkowitz, R. J. Seven-transmembrane-spanning receptors and heart function. Nature 415, 206–212 (2002).
Elphick, G. F. et al. The human polyomavirus, JCV, uses serotonin receptors to infect cells. Science 306, 1380–1383 (2004).
Roth, B. L. & Kroeze, W. K. Integrated approaches for genome-wide interrogation of the druggable non-olfactory G protein-coupled receptor superfamily. J. Biol. Chem. 290, 19471–19477 (2015).
Elkins, J. M. et al. Comprehensive characterization of the Published Kinase Inhibitor Set. Nat. Biotechnol. 34, 95–103 (2016).
Lin, X. et al. Life beyond kinases: structure-based discovery of sorafenib as nanomolar antagonist of 5-HT receptors. J. Med. Chem. 55, 5749–5759 (2012).
Huang, X.-P. et al. Allosteric ligands for the pharmacologically dark receptors GPR68 and GPR65. Nature 527, 477–483 (2015).
Chan, J. D. et al. The anthelmintic praziquantel is a human serotoninergic G-protein-coupled receptor ligand. Nat. Commun. 8, 1910 (2017).
Roth, B. L. Drugs and valvular heart disease. N. Engl. J. Med. 356, 6–9 (2007).
Kroeze, W. K. et al. PRESTO-Tango as an open-source resource for interrogation of the druggable human GPCRome. Nat. Struct. Mol. Biol. 22, 362–369 (2015).
Lansu, K. et al. In silico design of novel probes for the atypical opioid receptor MRGPRX2. Nat. Chem. Biol. 13, 529–536 (2017).
Pafilis, E. et al. The SPECIES and ORGANISMS Resources for Fast and Accurate Identification of Taxonomic Names in Text. PLoS ONE 8, e65390 (2013).
Okajima, D., Kudo, G. & Yokota, H. Antidepressant-like behavior in brain-specific angiogenesis inhibitor 2-deficient mice. J. Physiol. Sci. 61, 47–54 (2011).
Katsu, T. et al. The human frizzled-3 (FZD3) gene on chromosome 8p21, a receptor gene for Wnt ligands, is associated with the susceptibility to schizophrenia. Neurosci. Lett. 353, 53–56 (2003).
Wei, J. & Hemmings, G. P. Lack of a genetic association between the frizzled-3 gene and schizophrenia in a British population. Neurosci. Lett. 366, 336–338 (2004).
Jeong, S. H., Joo, E. J., Ahn, Y. M., Lee, K. Y. & Kim, Y. S. Investigation of genetic association between human Frizzled homolog 3 gene (FZD3) and schizophrenia: results in a Korean population and evidence from meta-analysis. Psychiatry Res. 143, 1–11 (2006).
Wu, P., Nielsen, T. E. & Clausen, M. H. Small-molecule kinase inhibitors: an analysis of FDA-approved drugs. Drug Discov. Today 21, 5–10 (2016).
Zawistowski, J. S. et al. Enhancer remodeling during adaptive bypass to MEK inhibition is attenuated by pharmacologic targeting of the P-TEFb complex. Cancer Discov. 7, 302–321 (2017).
Kullmann, D. M. The neuronal channelopathies. Brain 125, 1177–1195 (2002).
Gloyn, A. L. et al. Large-scale association studies of variants in genes encoding the pancreatic beta-cell KATP channel subunits Kir6.2 (KCNJ11) and SUR1 (ABCC8) confirm that the KCNJ11 E23K variant is associated with type 2 diabetes. Diabetes 52, 568–572 (2003).
Marbán, E. Cardiac channelopathies. Nature 415, 213–218 (2002).
Berman, R. M. et al. Antidepressant effects of ketamine in depressed patients. Biol. Psychiatry 47, 351–354 (2000).
Kirby, T. Ketamine for depression: the highs and lows. Lancet Psychiatry 2, 783–784 (2015).
Zanos, P. et al. NMDAR inhibition-independent antidepressant actions of ketamine metabolites. Nature 533, 481–486 (2016).
Pedersen, S. F., Klausen, T. K. & Nilius, B. The identification of a volume-regulated anion channel: an amazing Odyssey. Acta Physiol. 213, 868–881 (2015).
Niemeyer, B. A. Changing calcium: CRAC channel (STIM and Orai) expression, splicing, and posttranslational modifiers. Am. J. Physiol. Cell Physiol. 310, C701–709 (2016).
Dauner, K., Lissmann, J., Jeridi, S., Frings, S. & Möhrlen, F. Expression patterns of anoctamin 1 and anoctamin 2 chloride channels in the mammalian nose. Cell Tissue Res. 347, 327–341 (2012).
Pandey, A. K., Lu, L., Wang, X., Homayouni, R. & Williams, R. W. Functionally enigmatic genes: a case study of the brain ignorome. PLoS ONE 9, e88889 (2014).
Pfeffer, C. & Olsen, B. R. Editorial: Journal of negative results in biomedicine. J. Negat. Results Biomed. 1, 2 (2002).
Groth, P., Gibson, A. & Velterop, J. The anatomy of a nanopublication. Inf. Serv. Use 30, 51–56 (2010).
Agarwal, P. & Searls, D. B. Can literature analysis identify innovation drivers in drug discovery? Nat. Rev. Drug Discov. 8, 865–878 (2009).
Nguyen, D.-T. et al. Pharos: Collating protein information to shed light on the druggable genome. Nucleic Acids Res. 45, D995–D1002 (2017).
Wishart, D. S. et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 46, D1074–D1082 (2017).
The UniProt Consortium. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 45, D158–D169 (2017).
Griffith, M. et al. CIViC is a community knowledgebase for expert crowdsourcing the clinical interpretation of variants in cancer. Nat. Genet. 49, 170–174 (2017).
Koscielny, G. et al. Open Targets: a platform for therapeutic target identification and validation. Nucleic Acids Res. 45, D985–D994 (2017).
Lin, Y. et al. Drug target ontology to classify and integrate drug discovery data. J. Biomed. Semant. 8, 50 (2017).
Maggon, K. Best-selling human medicines 2002–2004. Drug Discov. Today 10, 739–742 (2005).
Stebbins, S. The world's 15 top selling drugs. 24/7 Wall St. http://247wallst.com/special-report/2016/04/26/top-selling-drugs-in-the-world/, (2016).
Hauser, A. S., Attwood, M. M., Rask-Andersen, M., Schiöth, H. B. & Gloriam, D. E. Trends in GPCR drug discovery: new agents, targets and indications. Nat. Rev. Drug Discov. 16, 829–842 (2017).
Shih, H.-P., Zhang, X. & Aronov, A. M. Drug discovery effectiveness from the standpoint of therapeutic mechanisms and indications. Nat. Rev. Drug Discov. 17, 19–33 (2018).
Tartaglia, L. A. et al. Identification and expression cloning of a leptin receptor, OB-R. Cell 83, 1263–1271 (1995).
Xie, J. et al. Activating Smoothened mutations in sporadic basal-cell carcinoma. Nature 391, 90–92 (1998).
Lee, M. J. et al. Sphingosine-1-phosphate as a ligand for the G protein-coupled receptor EDG-1. Science 279, 1552–1555 (1998).
Sakurai, T. et al. Orexins and orexin receptors: a family of hypothalamic neuropeptides and G protein-coupled receptors that regulate feeding behavior. Cell 92, 573–585 (1998).
Abifadel, M. et al. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat. Genet. 34, 154–156 (2003).
Kojima, M. et al. Ghrelin is a growth-hormone-releasing acylated peptide from stomach. Nature 402, 656–660 (1999).
Temel, J. S. et al. Anamorelin in patients with non-small-cell lung cancer and cachexia (ROMANA 1 and ROMANA 2): results from two randomised, double-blind, phase 3 trials. Lancet Oncol. 17, 519–531 (2016).
Acknowledgements
This work was supported by US National Institutes of Health (NIH) grants U54 CA189205 and U24 224370 (Illuminating the Druggable Genome Knowledge Management Center (IDG KMC)) at the University of New Mexico, Novo Nordisk Foundation Center for Protein Research, European Bioinformatics Institute (EBI) and University of Miami, U54 CA189201 and U24 CA224260 (A.M., Mount Sinai), P30 CA118100 (T.I.O., G.N.G. and L.A.S., UNM) and UL1 TR001449 (T.I.O. and L.A.S.), UM1 HG006370 (International Mouse Phenotyping Consortium, T.F.M. and I.T.), U01 MH104974 (B.L.R.), U01 MH104984 (S.T.), U01 MH105028 (M.T.M.), U01 MH105026 (J.Q. and A.M., Baylor) and U01 MH104999, R01 CA177993 and U24 DK116204 (S.G. and G.L.J.) and by the European Molecular Biology Laboratory (EMBL) and Wellcome Trust Strategic Awards WT086151/Z/08/Z and WT104104/Z/14/Z (A.G., A.H., A.R.L., A.K., J.P.O., and G.P.); and by Novo Nordisk Foundation Denmark grant NNF14CC0001 (S.B., L.J.L. and D.W). R.G., A.J., D.T.N., A.S., N.S., and G.Z.K. were supported by the Intramural Research Program, National Center for Advancing Translational Sciences (NCATS) and by U54 CA189205. Dedicated to Francisc Schneider (1933–2017).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
S.B. and L.J.J. are co-founders, shareholders and scientific advisory board members of Intomics A/S, an omics data integration company. A.C. and C.R. are employees of IQVIA, a company serving the combined industries of health information technologies and clinical research. I.T. is a current employee of Google Germany. G.P. is a current employee of GlaxoSmithKline, a global health-care company. D.M. is a current employee of AstraZeneca, a global, research-based biopharmaceutical company. J.P.O. is currently an employee of Medicines Discovery Catapult, a UK government-funded facility for collaborative research and development.
Related links
RELATED LINKS
Supplementary information
Supplementary Box S1
Context, time and knowledge management (PDF 190 kb)
Supplementary File S2
Plot of bioactivity values for major target classes. (PDF 675 kb)
Supplementary Information
Supplementary S3 table (XLSX 12754 kb)
Supplementary S4 table
Statistical significance results for the four validating metrics shown in Figure 2a (PDF 117 kb)
Supplementary S5 Box
Spotlight on the ionizing radiation proteome (PDF 301 kb)
Supplementary S7 table
Spotlight on selectivity (PDF 132 kb)
Supplementary information
Supplementary S8 table (XLSX 321 kb)
Supplementary information
Supplementary S9 table (XLSX 1011 kb)
Supplementary information
Supplementary S10 table (XLSX 15 kb)
Supplementary information
Supplementary S11 table (XLSX 111 kb)
Supplementary File S11
IMS Health (MIDAS) global drug sales data (2011-2015), organized by ATC level 2 codes and by protein class, normalized to percentage values. (PDF 1026 kb)
Glossary
- Drug
-
Externally administered, possibly endogenous but mostly xenobiotic, substances that are administered to patients in order to influence the outcome of a disease, syndrome or condition.
- Drug targets
-
Molecular entities present in living systems that, upon interaction with therapeutic agents or their by-products, result in modified biological responses that lead to therapeutic outcomes. The interaction between a drug and its target leads, directly or indirectly, to observable clinical outcomes.
- Druggable genome
-
Originally defined by Hopkins and Groom as the set of genes that encode proteins that could be modulated by an orally administered small molecule, as estimated by Lipinski's 'rule of five' guidelines.
- Mode of action
-
Referred to as 'mechanism of action' when the molecular interactions are well understood; describes the way in which drugs exert their intended therapeutic action, resulting in the intended therapeutic outcome.
Rights and permissions
About this article
Cite this article
Oprea, T., Bologa, C., Brunak, S. et al. Unexplored therapeutic opportunities in the human genome. Nat Rev Drug Discov 17, 317–332 (2018). https://doi.org/10.1038/nrd.2018.14
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrd.2018.14
This article is cited by
-
Encapsulation of HaCaT Secretome for Enhanced Wound Healing Capacity on Human Dermal Fibroblasts
Molecular Biotechnology (2024)
-
A small-molecule TNIK inhibitor targets fibrosis in preclinical and clinical models
Nature Biotechnology (2024)
-
About the dark corners in the gene function space of Escherichia coli remaining without illumination by scientific literature
Biology Direct (2023)
-
Calibrated geometric deep learning improves kinase–drug binding predictions
Nature Machine Intelligence (2023)
-
A preclinical secondary pharmacology resource illuminates target-adverse drug reaction associations of marketed drugs
Nature Communications (2023)